Quake: quality-aware detection and correction of sequencing errors
- PMID: 21114842
- PMCID: PMC3156955
- DOI: 10.1186/gb-2010-11-11-r116
Quake: quality-aware detection and correction of sequencing errors
Abstract
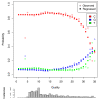
We introduce Quake, a program to detect and correct errors in DNA sequencing reads. Using a maximum likelihood approach incorporating quality values and nucleotide specific miscall rates, Quake achieves the highest accuracy on realistically simulated reads. We further demonstrate substantial improvements in de novo assembly and SNP detection after using Quake. Quake can be used for any size project, including more than one billion human reads, and is freely available as open source software from http://www.cbcb.umd.edu/software/quake.
© 2010 Kelley et al.; licensee BioMed Central Ltd.
Figures





References
-
- Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. - PubMed
-
- Haussler D, O'Brien S, Ryder O, Barker F, Clamp M, Crawford A, Hanner R, Hanotte O, Johnson W, McGuire J, Miller W, Murphy R, Murphy W, Sheldon F, Sinervo B, Venkatesh B, Wiley E, Allendorf F, Amato G, Baker C, Bauer A, Beja-Pereira A, Bermingham E, Bernardi G, Bonvicino C, Brenner S, Burke T, Cracraft J, Diekhans M, Edwards S. et al.Genome 10K: a proposal to obtain whole-genome sequence for 10 000 vertebrate species. J Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

