Comparative kinetics of Escherichia coli- and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha
- PMID: 21115717
- PMCID: PMC3028868
- DOI: 10.1128/IAI.01071-10
Comparative kinetics of Escherichia coli- and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha
Abstract
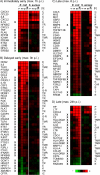
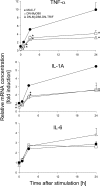
Infections of the udder by Escherichia coli very often elicit acute inflammation, while Staphylococcus aureus infections tend to cause mild, subclinical inflammation and persistent infections. The molecular causes underlying the different disease patterns are poorly understood. We therefore profiled the kinetics and extents of global changes in the transcriptome of primary bovine mammary epithelial cells (MEC) after challenging them with heat-inactivated preparations of E. coli or S. aureus pathogens. E. coli swiftly and strongly induced an expression of cytokines and bactericidal factors. S. aureus elicited a retarded response and failed to quickly induce an expression of bactericidal factors. Both pathogens induced similar patterns of chemokines for cell recruitment into the udder, but E. coli stimulated their synthesis much faster and stronger. The genes that are exclusively and most strongly upregulated by E. coli may be clustered into a regulatory network with tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1) in a central position. In contrast, the expression of these master cytokines is barely regulated by S. aureus. Both pathogens quickly trigger an enhanced expression of IL-6. This is still possible after completely abrogating MyD88-dependent Toll-like receptor (TLR) signaling in MEC. The E. coli-specific strong induction of TNF-α and IL-1 expression may be causative for the severe inflammatory symptoms of animals suffering from E. coli mastitis, while the avoidance to quickly induce the synthesis of bactericidal factors may support the persistent survival of S. aureus within the udder. We suggest that S. aureus subverts the MyD88-dependent activation of immune gene expression in MEC.
Figures



References
-
- Bannerman, D. D. 2009. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J. Anim. Sci. 87:10-25. - PubMed
-
- Bannerman, D. D., et al. 2004. Innate immune response to intramammary infection with Serratia marcescens and Streptococcus uberis. Vet. Res. 35:681-700. - PubMed
-
- Bannerman, D. D., M. J. Paape, W. R. Hare, and J. C. Hope. 2004. Characterization of the bovine innate immune response to intramammary infection with Klebsiella pneumoniae. J. Dairy Sci. 87:2420-2432. - PubMed
-
- Bannerman, D. D., M. J. Paape, and A. Chockalingam. 2006. Staphylococcus aureus intramammary infection elicits increased production of transforming growth factor-α, β1, and β2. Vet. Immunol. Immunopathol. 112:309-315. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

