Small RNA sorting: matchmaking for Argonautes
- PMID: 21116305
- PMCID: PMC3703915
- DOI: 10.1038/nrg2916
Small RNA sorting: matchmaking for Argonautes
Abstract
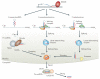
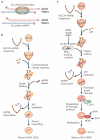
Small RNAs directly or indirectly impact nearly every biological process in eukaryotic cells. To perform their myriad roles, not only must precise small RNA species be generated, but they must also be loaded into specific effector complexes called RNA-induced silencing complexes (RISCs). Argonaute proteins form the core of RISCs and different members of this large family have specific expression patterns, protein binding partners and biochemical capabilities. In this Review, we explore the mechanisms that pair specific small RNA strands with their partner proteins, with an eye towards the substantial progress that has been recently made in understanding the sorting of the major small RNA classes - microRNAs (miRNAs) and small interfering RNAs (siRNAs) - in plants and animals.
Figures

References
-
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nature Rev. Genet. 2007;8:884–896. - PubMed
-
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. - PubMed
-
- Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. - PubMed
-
- Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

