The Viking viewer for connectomics: scalable multi-user annotation and summarization of large volume data sets
- PMID: 21118201
- PMCID: PMC3017751
- DOI: 10.1111/j.1365-2818.2010.03402.x
The Viking viewer for connectomics: scalable multi-user annotation and summarization of large volume data sets
Abstract
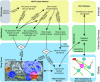
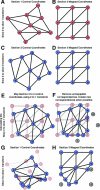
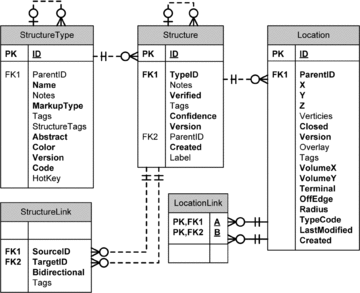
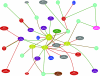
Modern microscope automation permits the collection of vast amounts of continuous anatomical imagery in both two and three dimensions. These large data sets present significant challenges for data storage, access, viewing, annotation and analysis. The cost and overhead of collecting and storing the data can be extremely high. Large data sets quickly exceed an individual's capability for timely analysis and present challenges in efficiently applying transforms, if needed. Finally annotated anatomical data sets can represent a significant investment of resources and should be easily accessible to the scientific community. The Viking application was our solution created to view and annotate a 16.5 TB ultrastructural retinal connectome volume and we demonstrate its utility in reconstructing neural networks for a distinctive retinal amacrine cell class. Viking has several key features. (1) It works over the internet using HTTP and supports many concurrent users limited only by hardware. (2) It supports a multi-user, collaborative annotation strategy. (3) It cleanly demarcates viewing and analysis from data collection and hosting. (4) It is capable of applying transformations in real-time. (5) It has an easily extensible user interface, allowing addition of specialized modules without rewriting the viewer.
© 2010 The Authors Journal of Microscopy © 2010 The Royal Microscopical Society.
Figures




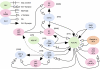

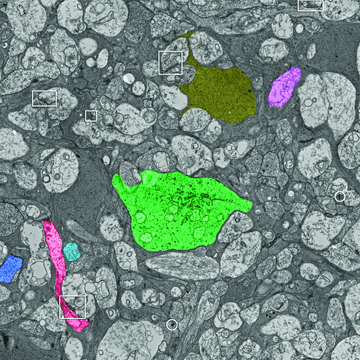
References
-
- Amari SI, Beltrame F, Bjaalie JG, et al. Neuroinformatics: the integration of shared databases and tools towards integrative neuroscience. J. Integr. Neurosci. 2002;1:117–128. - PubMed
-
- Fiala JC. Reconstruct: a free editor for serial section microscopy. J. Microsc. 2005;218:52–61. - PubMed
-
- Gansner ER, North SC. An open graph visualization system and its applications to software engineering. Softw. – Pract. Exp. 1999;30:1203–1233.
-
- Google. KML Reference – KML – Google Code. Google, Inc; 2010.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

