H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo
- PMID: 21118997
- PMCID: PMC3020919
- DOI: 10.1091/mbc.E10-07-0633
H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo
Abstract
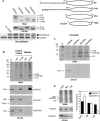
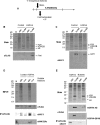
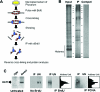
While specific posttranslational modification patterns within the H3 and H4 tail domains are associated with the S-phase, their actual functions in replication-dependent chromatin assembly have not yet been defined. Here we used incorporation of trace amounts of recombinant proteins into naturally synchronous macroplasmodia of Physarum polycephalum to examine the function of H3 and H4 tail domains in replication-coupled chromatin assembly. We found that the H3/H4 complex lacking the H4 tail domain was not efficiently recovered in nuclei, whereas depletion of the H3 tail domain did not impede nuclear import but chromatin assembly failed. Furthermore, our results revealed that the proper pattern of acetylation on the H4 tail domain is required for nuclear import and chromatin assembly. This is most likely due to binding of Hat1, as coimmunoprecipitation experiments showed Hat1 associated with predeposition histones in the cytoplasm and with replicating chromatin. These results suggest that the type B histone acetyltransferase assists in shuttling the H3/H4 complex from cytoplasm to the replication forks.
Figures



References
-
- Ahmad A, Takami Y, Nakayama T. Distinct regions of the chicken p46 polypeptide are required for its in vitro interaction with histones H2B and H4 and histone acetyltransferase-1. Biochem Biophys Res Commun. 2000;279:95–102. - PubMed
-
- Ai X, Parthun MR. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell. 2004;14:195–205. - PubMed
-
- Barman HK, Takami Y, Ono T, Nishijima H, Sanematsu F, Shibahara K, Nakayama T. Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem Biophys Res Commun. 2006;345:1547–1557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

