Folded DNA in action: hairpin formation and biological functions in prokaryotes
- PMID: 21119018
- PMCID: PMC3008174
- DOI: 10.1128/MMBR.00026-10
Folded DNA in action: hairpin formation and biological functions in prokaryotes
Abstract
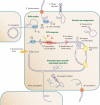
Structured forms of DNA with intrastrand pairing are generated in several cellular processes and are involved in biological functions. These structures may arise on single-stranded DNA (ssDNA) produced during replication, bacterial conjugation, natural transformation, or viral infections. Furthermore, negatively supercoiled DNA can extrude inverted repeats as hairpins in structures called cruciforms. Whether they are on ssDNA or as cruciforms, hairpins can modify the access of proteins to DNA, and in some cases, they can be directly recognized by proteins. Folded DNAs have been found to play an important role in replication, transcription regulation, and recognition of the origins of transfer in conjugative elements. More recently, they were shown to be used as recombination sites. Many of these functions are found on mobile genetic elements likely to be single stranded, including viruses, plasmids, transposons, and integrons, thus giving some clues as to the manner in which they might have evolved. We review here, with special focus on prokaryotes, the functions in which DNA secondary structures play a role and the cellular processes giving rise to them. Finally, we attempt to shed light on the selective pressures leading to the acquisition of functions for DNA secondary structures.
Figures
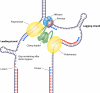




References
-
- Adhya, S. 1989. Multipartite genetic control elements: communication by DNA loop. Annu. Rev. Genet. 23:227-250. - PubMed
-
- Aleshkin, G. I., K. V. Kadzhaev, and A. P. Markov. 1998. High and low UV-dose responses in SOS-induction of the precise excision of transposons Tn1, Tn5 and Tn10 in Escherichia coli. Mutat. Res. 401:179-191. - PubMed
-
- Althorpe, N. J., P. M. Chilley, A. T. Thomas, W. J. Brammar, and B. M. Wilkins. 1999. Transient transcriptional activation of the Incl1 plasmid anti-restriction gene (ardA) and SOS inhibition gene (psiB) early in conjugating recipient bacteria. Mol. Microbiol. 31:133-142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

