Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase
- PMID: 21121604
- PMCID: PMC3076064
- DOI: 10.1021/ja1087612
Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase
Abstract
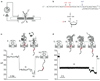
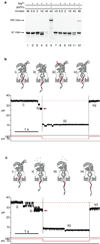
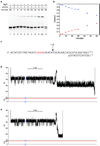
Coupling nucleic acid processing enzymes to nanoscale pores allows controlled movement of individual DNA or RNA strands that is reported as an ionic current/time series. Hundreds of individual enzyme complexes can be examined in single-file order at high bandwidth and spatial resolution. The bacteriophage phi29 DNA polymerase (phi29 DNAP) is an attractive candidate for this technology, due to its remarkable processivity and high affinity for DNA substrates. Here we show that phi29 DNAP-DNA complexes are stable when captured in an electric field across the α-hemolysin nanopore. DNA substrates were activated for replication at the nanopore orifice by exploiting the 3'-5' exonuclease activity of wild-type phi29 DNAP to excise a 3'-H terminal residue, yielding a primer strand 3'-OH. In the presence of deoxynucleoside triphosphates, DNA synthesis was initiated, allowing real-time detection of numerous sequential nucleotide additions that was limited only by DNA template length. Translocation of phi29 DNAP along DNA substrates was observed in real time at Ångstrom-scale precision as the template strand was drawn through the nanopore lumen during replication.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

