Identification and characterization of a bacterial glutamic peptidase
- PMID: 21122090
- PMCID: PMC3009609
- DOI: 10.1186/1471-2091-11-47
Identification and characterization of a bacterial glutamic peptidase
Abstract
Background: Glutamic peptidases, from the MEROPS family G1, are a distinct group of peptidases characterized by a catalytic dyad consisting of a glutamate and a glutamine residue, optimal activity at acidic pH and insensitivity towards the microbial derived protease inhibitor, pepstatin. Previously, only glutamic peptidases derived from filamentous fungi have been characterized.
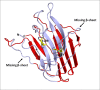
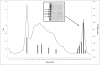
Results: We report the first characterization of a bacterial glutamic peptidase (pepG1), derived from the thermoacidophilic bacteria Alicyclobacillus sp. DSM 15716. The amino acid sequence identity between pepG1 and known fungal glutamic peptidases is only 24-30% but homology modeling, the presence of the glutamate/glutamine catalytic dyad and a number of highly conserved motifs strongly support the inclusion of pepG1 as a glutamic peptidase. Phylogenetic analysis places pepG1 and other putative bacterial and archaeal glutamic peptidases in a cluster separate from the fungal glutamic peptidases, indicating a divergent and independent evolution of bacterial and fungal glutamic peptidases. Purification of pepG1, heterologously expressed in Bacillus subtilis, was performed using hydrophobic interaction chromatography and ion exchange chromatography. The purified peptidase was characterized with respect to its physical properties. Temperature and pH optimums were found to be 60°C and pH 3-4, in agreement with the values observed for the fungal members of family G1. In addition, pepG1 was found to be pepstatin-insensitive, a characteristic signature of glutamic peptidases.
Conclusions: Based on the obtained results, we suggest that pepG1 can be added to the MEROPS family G1 as the first characterized bacterial member.
Figures



References
-
- Becker F, Schnorr K, Wilting R, Tolstrup N, Bendtsen JD, Olsen PB. Development of in vitro transposon assisted signal sequence trapping and its use in screening Bacillus halodurans C125 and Sulfolobus solfataricus P2 gene libraries. J Microbiol Methods. 2004;57(1):123–133. doi: 10.1016/j.mimet.2003.12.002. - DOI - PubMed
-
- Inoue H, Kimura T, Makabe O, Takahashi K. The gene and deduced protein sequences of the zymogen of Aspergillus niger acid proteinase A. J Biol Chem. 1991;266(29):19484–19489. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

