Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer
- PMID: 21122106
- PMCID: PMC3046450
- DOI: 10.1186/bcr2788
Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer
Abstract
Introduction: Mutational inactivation of the FBXW7/hCDC4 tumor suppressor gene (TSG) is common in many cancer types, but infrequent in breast cancers. This study investigates the presence and impact of FBXW7/hCDC4 promoter methylation in breast cancer.
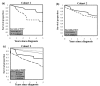
Methods: FBXW7/hCDC4-β expression and promoter methylation was assessed in 161 tumors from two independent breast cancer cohorts. Associations between methylation status and clinicopathologic characteristics were assessed by Fisher's exact test. Survival was analyzed using the Kaplan-Meier method in addition to modeling the risk by use of a multivariate proportional hazard (Cox) model adjusting for possible confounders of survival.
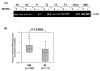
Results: Methylation of the promoter and loss of mRNA expression was found both in cell lines and primary tumors (43% and 51%, respectively). Using Cox modeling, a trend was found towards decreased hazard ratio (HR) for death in women with methylation of FBXW7/hCDC4-β in both cohorts (HR 0.53 (95% CI 0.23 to 1.23) and HR 0.50 (95% CI 0.23 to 1.08), respectively), despite an association between methylation and high-grade tumors (P = 0.017). Interestingly, in subgroups of patients whose tumors are p53 mutated or lymph-node positive, promoter methylation identified patients with significantly improved survival (P = 0.048 and P = 0.017, respectively).
Conclusions: We demonstrate an alternative mechanism for inactivation of the TSG FBXW7/hCDC4, namely promoter specific methylation. Importantly, in breast cancer, methylation of FBXW7/hCDC4-β is related to favorable prognosis despite its association with poorly differentiated tumors. Future work may define whether FBXW7/hCDC4 methylation is a biomarker of the response to chemotherapy and a target for epigenetic modulation therapy.
Figures


Similar articles
-
FBXW7 is involved in the acquisition of the malignant phenotype in epithelial ovarian tumors.Cancer Sci. 2016 Oct;107(10):1399-1405. doi: 10.1111/cas.13026. Epub 2016 Sep 24. Cancer Sci. 2016. PMID: 27486687 Free PMC article.
-
Promoter hypermethylation is not the major mechanism for inactivation of the FBXW7 beta-form in human gliomas.Genes Genet Syst. 2008 Aug;83(4):347-52. doi: 10.1266/ggs.83.347. Genes Genet Syst. 2008. PMID: 18931460
-
The Fbxw7/hCdc4 tumor suppressor in human cancer.Cancer Lett. 2008 Nov 18;271(1):1-12. doi: 10.1016/j.canlet.2008.04.036. Epub 2008 Jun 9. Cancer Lett. 2008. PMID: 18541364 Review.
-
Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer.Cancer Res. 2004 Feb 1;64(3):795-800. doi: 10.1158/0008-5472.can-03-3417. Cancer Res. 2004. PMID: 14871801
-
The Role of FBXW7 in Gynecologic Malignancies.Cells. 2023 May 17;12(10):1415. doi: 10.3390/cells12101415. Cells. 2023. PMID: 37408248 Free PMC article. Review.
Cited by
-
Roles of F-box proteins in cancer.Nat Rev Cancer. 2014 Apr;14(4):233-47. doi: 10.1038/nrc3700. Nat Rev Cancer. 2014. PMID: 24658274 Free PMC article. Review.
-
A novel peptide 66CTG stabilizes Myc proto-oncogene protein to promote triple-negative breast cancer growth.Signal Transduct Target Ther. 2025 Jul 9;10(1):217. doi: 10.1038/s41392-025-02298-5. Signal Transduct Target Ther. 2025. PMID: 40628713 Free PMC article.
-
FBXW7 regulates the sensitivity of imatinib in gastrointestinal stromal tumors by targeting MCL1.Gastric Cancer. 2024 Mar;27(2):235-247. doi: 10.1007/s10120-023-01454-6. Epub 2023 Dec 24. Gastric Cancer. 2024. PMID: 38142463
-
Loss of FBXW7 and accumulation of MCL1 and PLK1 promote paclitaxel resistance in breast cancer.Oncotarget. 2016 Aug 16;7(33):52751-52765. doi: 10.18632/oncotarget.10481. Oncotarget. 2016. PMID: 27409838 Free PMC article.
-
Overexpression of F-box and WD repeat domain containing 7 prevents tumor growth of bladder cancer cells through regulating SREBP1a.Transl Androl Urol. 2022 Mar;11(3):367-376. doi: 10.21037/tau-22-146. Transl Androl Urol. 2022. PMID: 35402194 Free PMC article.
References
-
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, Mueller-Holzner E, Corcoran M, Dagnell M, Nejad SZ, Nayer BN, Zali MR, Hansson J, Egyhazi S, Petersson F, Sangfelt P, Nordgren H, Grander D, Reed SI, Widschwendter M, Sangfelt O, Spruck C. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

