Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs
- PMID: 21123178
- PMCID: PMC3037645
- DOI: 10.1074/jbc.M110.176339
Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs
Abstract
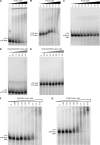
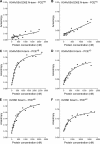
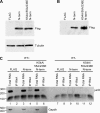
The DExH protein RNA helicase A (RHA) plays numerous roles in cell physiology, and post-transcriptional activation of gene expression is a major role among them. RHA selectively activates translation of complex cellular and retroviral mRNAs. Although RHA requires interaction with structural features of the 5'-UTR of these target mRNAs, the molecular basis of their translation activation by RHA is poorly understood. RHA contains a conserved ATPase-dependent helicase core that is flanked by two α-β-β-β-α double-stranded RNA-binding domains at the N terminus and repeated arginine-glycine residues at the C terminus. The individual recombinant N-terminal, central helicase, and C-terminal domains were evaluated for their ability to specifically interact with cognate RNAs by in vitro biochemical measurements and mRNA translation assays in cells. The results demonstrate that N-terminal residues confer selective interaction with retroviral and junD target RNAs. Conserved lysine residues in the distal α-helix of the double-stranded RNA-binding domains are necessary to engage structural features of retroviral and junD 5'-UTRs. Exogenous expression of the N terminus coprecipitates junD mRNA and inhibits the translation activity of endogenous RHA. The results indicate that the molecular basis for the activation of translation by RHA is recognition of target mRNA by the N-terminal domain that tethers the ATP-dependent helicase for rearrangement of the complex 5'-UTR.
Figures





Similar articles
-
A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs.RNA. 2012 Feb;18(2):321-34. doi: 10.1261/rna.030288.111. Epub 2011 Dec 21. RNA. 2012. PMID: 22190748 Free PMC article.
-
Different activities of the conserved lysine residues in the double-stranded RNA binding domains of RNA helicase A in vitro and in the cell.Biochim Biophys Acta. 2014 Jul;1840(7):2234-43. doi: 10.1016/j.bbagen.2014.04.003. Epub 2014 Apr 12. Biochim Biophys Acta. 2014. PMID: 24726449
-
RNA helicase A is necessary for translation of selected messenger RNAs.Nat Struct Mol Biol. 2006 Jun;13(6):509-16. doi: 10.1038/nsmb1092. Epub 2006 May 7. Nat Struct Mol Biol. 2006. PMID: 16680162
-
The Current View on the Helicase Activity of RNA Helicase A and Its Role in Gene Expression.Curr Protein Pept Sci. 2021;22(1):29-40. doi: 10.2174/1389203721666201103084122. Curr Protein Pept Sci. 2021. PMID: 33143622 Review.
-
RNA Helicase A Regulates the Replication of RNA Viruses.Viruses. 2021 Feb 25;13(3):361. doi: 10.3390/v13030361. Viruses. 2021. PMID: 33668948 Free PMC article. Review.
Cited by
-
In vitro and in vivo analysis of the interaction between RNA helicase A and HIV-1 RNA.J Virol. 2012 Dec;86(24):13272-80. doi: 10.1128/JVI.01993-12. Epub 2012 Sep 26. J Virol. 2012. PMID: 23015696 Free PMC article.
-
Cell type-dependent gene regulation by Staufen2 in conjunction with Upf1.BMC Mol Biol. 2011 Nov 16;12:48. doi: 10.1186/1471-2199-12-48. BMC Mol Biol. 2011. PMID: 22087843 Free PMC article.
-
Isolation of Cognate RNA-protein Complexes from Cells Using Oligonucleotide-directed Elution.J Vis Exp. 2017 Jan 16;(119):54391. doi: 10.3791/54391. J Vis Exp. 2017. PMID: 28117770 Free PMC article.
-
Generation of Dhx9-deficient clones in T-cell development with a mitotic recombination technique.Genesis. 2012 Jul;50(7):543-51. doi: 10.1002/dvg.22005. Genesis. 2012. PMID: 22988576 Free PMC article.
-
The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.Nucleic Acids Res. 2021 Jun 4;49(10):5925-5942. doi: 10.1093/nar/gkab342. Nucleic Acids Res. 2021. PMID: 33978756 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

