Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants
- PMID: 21123529
- PMCID: PMC3043520
- DOI: 10.1128/JCM.01588-10
Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants
Abstract
Carbapenem resistance mediated by plasmid-borne Klebsiella pneumoniae carbapenemases (KPC) is an emerging problem of significant clinical importance in Gram-negative bacteria. Multiple KPC gene variants (bla(KPC)) have been reported, with KPC-2 (bla(KPC-2)) and KPC-3 (bla(KPC-3)) associated with epidemic outbreaks in New York City and various international settings. Here, we describe the development of a multiplex real-time PCR assay using molecular beacons (MB-PCR) for rapid and accurate identification of bla(KPC) variants. The assay consists of six molecular beacons and two oligonucleotide primer pairs, allowing for detection and classification of all currently described bla(KPC) variants (bla(KPC-2) to bla(KPC-11)). The MB-PCR detection limit was 5 to 40 DNA copies per reaction and 4 CFU per reaction using laboratory-prepared samples. The MB-PCR probes were highly specific for each bla(KPC) variant, and cross-reactivity was not observed using DNA isolated from several bacterial species. A total of 457 clinical Gram-negative isolates were successfully characterized by our MB-PCR assay, with bla(KPC-3) and bla(KPC-2) identified as the most common types in the New York/New Jersey metropolitan region. The MB-PCR assay described herein is rapid, sensitive, and specific and should be useful for understanding the ongoing evolution of carbapenem resistance in Gram-negative bacteria. As novel bla(KPC) variants continue to emerge, the MB-PCR assay can be modified in response to epidemiologic developments.
Figures
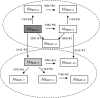
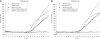
References
-
- Bradford P., et al. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55–60 - PubMed
-
- Bratu S., et al. 2007. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin. Infect. Dis. 44:972–975 - PubMed
-
- Bratu S., et al. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128–132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

