Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection
- PMID: 21124791
- PMCID: PMC2991352
- DOI: 10.1371/journal.pone.0014116
Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection
Abstract
Background: The microbiome of the male urogenital tract is poorly described but it has been suggested that bacterial colonization of the male urethra might impact risk of sexually transmitted infection (STI). Previous cultivation-dependent studies showed that a variety of non-pathogenic bacteria colonize the urethra but did not thoroughly characterize these microbiomes or establish links between the compositions of urethral microbiomes and STI.
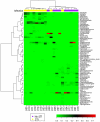
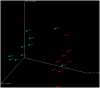
Methodology/findings: Here, we used 16S rRNA PCR and sequencing to identify bacteria in urine specimens collected from men who lacked symptoms of urethral inflammation but who differed in status for STI. All of the urine samples contained multiple bacterial genera and many contained taxa that colonize the human vagina. Uncultivated bacteria associated with female genital tract pathology were abundant in specimens from men who had STI.
Conclusions: Urine microbiomes from men with STI were dominated by fastidious, anaerobic and uncultivated bacteria. The same taxa were rare in STI negative individuals. Our findings suggest that the composition of male urine microbiomes is related to STI.
Conflict of interest statement
Figures

References
-
- Frank DN, Pace NR. Molecular-phylogenetic analyses of human gastrointestinal microbiota. Curr Opin Gastroenterol. 2001;17:52–57. - PubMed
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

