Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans
- PMID: 21124819
- PMCID: PMC2991264
- DOI: 10.1371/journal.pgen.1001219
Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans
Abstract
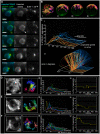
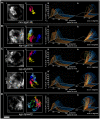
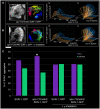
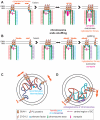
The Caenorhabditis elegans inner nuclear envelope protein matefin/SUN-1 plays a conserved, pivotal role in the process of genome haploidization. CHK-2-dependent phosphorylation of SUN-1 regulates homologous chromosome pairing and interhomolog recombination in Caenorhabditis elegans. Using time-lapse microscopy, we characterized the movement of matefin/SUN-1::GFP aggregates (the equivalent of chromosomal attachment plaques) and showed that the dynamics of matefin/SUN-1 aggregates remained unchanged throughout leptonene/zygotene, despite the progression of pairing. Movement of SUN-1 aggregates correlated with chromatin polarization. We also analyzed the requirements for the formation of movement-competent matefin/SUN-1 aggregates in the context of chromosome structure and found that chromosome axes were required to produce wild-type numbers of attachment plaques. Abrogation of synapsis led to a deceleration of SUN-1 aggregate movement. Analysis of matefin/SUN-1 in a double-strand break deficient mutant revealed that repair intermediates influenced matefin/SUN-1 aggregate dynamics. Investigation of movement in meiotic regulator mutants substantiated that proper orchestration of the meiotic program and effective repair of DNA double-strand breaks were necessary for the wild-type behavior of matefin/SUN-1 aggregates.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Petronczki M, Siomos M, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. - PubMed
-
- Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. - PubMed
-
- Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma. 2007;116:497–505. - PubMed
-
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

