Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
- PMID: 21124847
- PMCID: PMC2990756
- DOI: 10.1371/journal.pone.0015535
Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Abstract
Background: Small molecule modulators of epigenetic processes are currently sought as basic probes for biochemical mechanisms, and as starting points for development of therapeutic agents. N(ε)-Methylation of lysine residues on histone tails is one of a number of post-translational modifications that together enable transcriptional regulation. Histone lysine demethylases antagonize the action of histone methyltransferases in a site- and methylation state-specific manner. N(ε)-Methyllysine demethylases that use 2-oxoglutarate as co-factor are associated with diverse human diseases, including cancer, inflammation and X-linked mental retardation; they are proposed as targets for the therapeutic modulation of transcription. There are few reports on the identification of templates that are amenable to development as potent inhibitors in vivo and large diverse collections have yet to be exploited for the discovery of demethylase inhibitors.
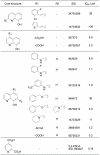
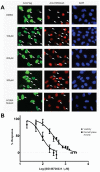
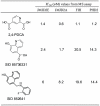
Principal findings: High-throughput screening of a ∼236,000-member collection of diverse molecules arrayed as dilution series was used to identify inhibitors of the JMJD2 (KDM4) family of 2-oxoglutarate-dependent histone demethylases. Initial screening hits were prioritized by a combination of cheminformatics, counterscreening using a coupled assay enzyme, and orthogonal confirmatory detection of inhibition by mass spectrometric assays. Follow-up studies were carried out on one of the series identified, 8-hydroxyquinolines, which were shown by crystallographic analyses to inhibit by binding to the active site Fe(II) and to modulate demethylation at the H3K9 locus in a cell-based assay.
Conclusions: These studies demonstrate that diverse compound screening can yield novel inhibitors of 2OG dependent histone demethylases and provide starting points for the development of potent and selective agents to interrogate epigenetic regulation.
Conflict of interest statement
Figures



References
-
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. - PubMed
-
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. - PubMed
-
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of Lysine 4 on Histone H3: Intricacy of Writing and Reading a Single Epigenetic Mark. Molecular Cell. 2007;25:15–30. - PubMed
-
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

