An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain
- PMID: 21124902
- PMCID: PMC2990719
- DOI: 10.1371/journal.pone.0014102
An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain
Abstract
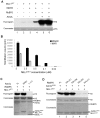
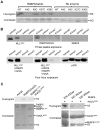
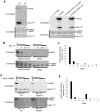
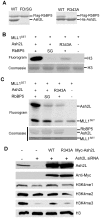
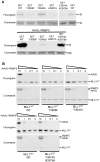
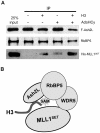
Histone H3 lysine 4 (K4) methylation is a prevalent mark associated with transcription activation and is mainly catalyzed by the MLL/SET1 family histone methyltransferases. A common feature of the mammalian MLL/SET1 complexes is the presence of three core components (RbBP5, Ash2L and WDR5) and a catalytic subunit containing a SET domain. Unlike most other histone lysine methyltransferases, all four proteins are required for efficient H3 K4 methylation. Despite extensive efforts, mechanisms for how three core components regulate MLL/SET1 methyltransferase activity remain elusive. Here we show that a heterodimer of Ash2L and RbBP5 has intrinsic histone methyltransferase activity. This activity requires the highly conserved SPRY domain of Ash2L and a short peptide of RbBP5. We demonstrate that both Ash2L and the MLL1 SET domain are capable of binding to S-adenosyl-L- [methyl-(3)H] methionine in the MLL1 core complex. Mutations in the MLL1 SET domain that fail to support overall H3 K4 methylation also compromise SAM binding by Ash2L. Taken together, our results show that the Ash2L/RbBP5 heterodimer plays a critical role in the overall catalysis of MLL1 mediated H3 K4 methylation. The results we describe here provide mechanistic insights for unique regulation of the MLL1 methyltransferase activity. It suggests that both Ash2L/RbBP5 and the MLL1 SET domain make direct contacts with the substrates and contribute to the formation of a joint catalytic center. Given the shared core configuration among all MLL/SET1 family HMTs, it will be interesting to test whether the mechanism we describe here can be generalized to other MLL/SET1 family members in the future.
Conflict of interest statement
Figures
References
-
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. - PubMed
-
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. - PubMed
-
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. - PubMed
-
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

