The preferred substrates for transglutaminase 2 in a complex wheat gluten digest are Peptide fragments harboring celiac disease T-cell epitopes
- PMID: 21124911
- PMCID: PMC2988817
- DOI: 10.1371/journal.pone.0014056
The preferred substrates for transglutaminase 2 in a complex wheat gluten digest are Peptide fragments harboring celiac disease T-cell epitopes
Abstract
Background: Celiac disease is a T-cell mediated chronic inflammatory disorder of the gut that is induced by dietary exposure to gluten proteins. CD4+ T cells of the intestinal lesion recognize gluten peptides in the context of HLA-DQ2.5 or HLA-DQ8 and the gluten derived peptides become better T-cell antigens after deamidation catalyzed by the enzyme transglutaminase 2 (TG2). In this study we aimed to identify the preferred peptide substrates of TG2 in a heterogeneous proteolytic digest of whole wheat gluten.
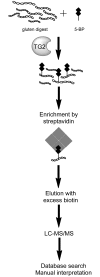
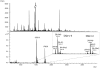
Methods: A method was established to enrich for preferred TG2 substrates in a complex gluten peptide mixture by tagging with 5-biotinamido-pentylamine. Tagged peptides were isolated and then identified by nano-liquid chromatography online-coupled to tandem mass spectrometry, database searching and final manual data validation.
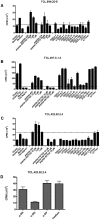
Results: We identified 31 different peptides as preferred substrates of TG2. Strikingly, the majority of these peptides were harboring known gluten T-cell epitopes. Five TG2 peptide substrates that were predicted to bind to HLA-DQ2.5 did not contain previously characterized sequences of T-cell epitopes. Two of these peptides elicited T-cell responses when tested for recognition by intestinal T-cell lines of celiac disease patients, and thus they contain novel candidate T-cell epitopes. We also found that the intact 9mer core sequences of the respective epitopes were not present in all peptide substrates. Interestingly, those epitopes that were represented by intact forms were frequently recognized by T cells in celiac disease patients, whereas those that were present in truncated versions were infrequently recognized.
Conclusion: TG2 as well as gastrointestinal proteolysis play important roles in the selection of gluten T-cell epitopes in celiac disease.
Conflict of interest statement
Figures
References
-
- Sollid LM. Molecular basis of celiac disease. Annu Rev Immunol. 2000;18:53–81. - PubMed
-
- Molberg O, Kett K, Scott H, Thorsby E, Sollid LM, et al. Gliadin specific, HLA DQ2-restricted T cells are commonly found in small intestinal biopsies from coeliac disease patients, but not from controls. Scand J Immunol. 1997;46:103–109. - PubMed
-
- Johansen BH, Vartdal F, Eriksen JA, Thorsby E, Sollid LM. Identification of a putative motif for binding of peptides to HLA-DQ2. Int Immunol. 1996;8:177–182. - PubMed
-
- van der Wal Y, Kooy YM, Drijfhout JW, Amons R, Koning F. Peptide binding characteristics of the coeliac disease-associated DQ(alpha1*0501, beta1*0201) molecule. Immunogenetics. 1996;44:246–253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

