Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists
- PMID: 21126361
- PMCID: PMC3014934
- DOI: 10.1186/1471-2148-10-377
Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists
Abstract
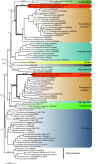
Background: Recent phylogenomic analyses have revolutionized our view of eukaryote evolution by revealing unexpected relationships between and within the eukaryotic supergroups. However, for several groups of uncultivable protists, only the ribosomal RNA genes and a handful of proteins are available, often leading to unresolved evolutionary relationships. A striking example concerns the supergroup Rhizaria, which comprises several groups of uncultivable free-living protists such as radiolarians, foraminiferans and gromiids, as well as the parasitic plasmodiophorids and haplosporids. Thus far, the relationships within this supergroup have been inferred almost exclusively from rRNA, actin, and polyubiquitin genes, and remain poorly resolved. To address this, we have generated large Expressed Sequence Tag (EST) datasets for 5 species of Rhizaria belonging to 3 important groups: Acantharea (Astrolonche sp., Phyllostaurus sp.), Phytomyxea (Spongospora subterranea, Plasmodiophora brassicae) and Gromiida (Gromia sphaerica).
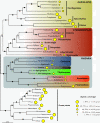
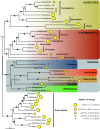
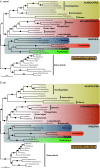
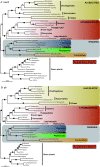
Results: 167 genes were selected for phylogenetic analyses based on the representation of at least one rhizarian species for each gene. Concatenation of these genes produced a supermatrix composed of 36,735 amino acid positions, including 10 rhizarians, 9 stramenopiles, and 9 alveolates. Phylogenomic analyses of this large dataset revealed a strongly supported clade grouping Foraminifera and Acantharea. The position of this clade within Rhizaria was sensitive to the method employed and the taxon sampling: Maximum Likelihood (ML) and Bayesian analyses using empirical model of evolution favoured an early divergence, whereas the CAT model and ML analyses with fast-evolving sites or the foraminiferan species Reticulomyxa filosa removed suggested a derived position, closely related to Gromia and Phytomyxea. In contrast to what has been previously reported, our analyses also uncovered the presence of the rhizarian-specific polyubiquitin insertion in Acantharea. Finally, this work reveals another possible rhizarian signature in the 60S ribosomal protein L10a.
Conclusions: Our study provides new insights into the evolution of Rhizaria based on phylogenomic analyses of ESTs from three groups of previously under-sampled protists. It was enabled through the application of a recently developed method of transcriptome analysis, requiring very small amount of starting material. Our study illustrates the potential of this method to elucidate the early evolution of eukaryotes by providing large amount of data for uncultivable free-living and parasitic protists.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

