Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes
- PMID: 21127052
- PMCID: PMC3037656
- DOI: 10.1074/jbc.M110.205054
Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes
Abstract
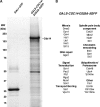
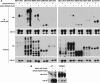
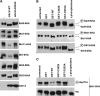
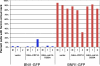
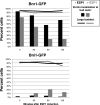
Cdc14 phosphatase regulates multiple events during anaphase and is essential for mitotic exit in budding yeast. Cdc14 is regulated in both a spatial and temporal manner. It is sequestered in the nucleolus for most of the cell cycle by the nucleolar protein Net1 and is released into the nucleus and cytoplasm during anaphase. To identify novel binding partners of Cdc14, we used affinity purification of Cdc14 and mass spectrometric analysis of interacting proteins from strains in which Cdc14 localization or catalytic activity was altered. To alter Cdc14 localization, we used a strain deleted for NET1, which causes full release of Cdc14 from the nucleolus. To alter Cdc14 activity, we generated mutations in the active site of Cdc14 (C283S or D253A), which allow binding of substrates, but not dephosphorylation, by Cdc14. Using this strategy, we identified new interactors of Cdc14, including multiple proteins involved in mitotic events. A subset of these proteins displayed increased affinity for catalytically inactive mutants of Cdc14 compared with the wild-type version, suggesting they are likely substrates of Cdc14. We have also shown that several of the novel Cdc14-interacting proteins, including Kar9 (a protein that orients the mitotic spindle) and Bni1 and Bnr1 (formins that nucleate actin cables and may be important for actomyosin ring contraction) are specifically dephosphorylated by Cdc14 in vitro and in vivo. Our findings suggest the dephosphorylation of the formins may be important for their observed localization change during exit from mitosis and indicate that Cdc14 targets proteins involved in wide-ranging mitotic events.
Figures




References
-
- Schwab M., Lutum A. S., Seufert W. (1997) Cell 90, 683–693 - PubMed
-
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. (1998) Mol. Cell 2, 709–718 - PubMed
-
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Shevchenko A., Charbonneau H., Deshaies R. J. (1999) Cell 97, 233–244 - PubMed
-
- Visintin R., Hwang E. S., Amon A. (1999) Nature 398, 818–823 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

