Rewiring of genetic networks in response to DNA damage
- PMID: 21127252
- PMCID: PMC3006187
- DOI: 10.1126/science.1195618
Rewiring of genetic networks in response to DNA damage
Erratum in
- Science. 2011 Jan 21;331(6015):284
Abstract
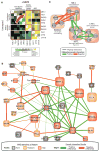
Although cellular behaviors are dynamic, the networks that govern these behaviors have been mapped primarily as static snapshots. Using an approach called differential epistasis mapping, we have discovered widespread changes in genetic interaction among yeast kinases, phosphatases, and transcription factors as the cell responds to DNA damage. Differential interactions uncover many gene functions that go undetected in static conditions. They are very effective at identifying DNA repair pathways, highlighting new damage-dependent roles for the Slt2 kinase, Pph3 phosphatase, and histone variant Htz1. The data also reveal that protein complexes are generally stable in response to perturbation, but the functional relations between these complexes are substantially reorganized. Differential networks chart a new type of genetic landscape that is invaluable for mapping cellular responses to stimuli.
Figures



Comment in
-
Genetics. The DNA damage road map.Science. 2010 Dec 3;330(6009):1327-8. doi: 10.1126/science.1199862. Science. 2010. PMID: 21127235 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

