Evolution of domain promiscuity in eukaryotic genomes--a perspective from the inferred ancestral domain architectures
- PMID: 21127809
- PMCID: PMC3321261
- DOI: 10.1039/c0mb00182a
Evolution of domain promiscuity in eukaryotic genomes--a perspective from the inferred ancestral domain architectures
Abstract
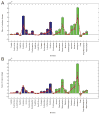
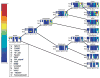
Most eukaryotic proteins are composed of two or more domains. These assemble in a modular manner to create new proteins usually by the acquisition of one or more domains to an existing protein. Promiscuous domains which are found embedded in a variety of proteins and co-exist with many other domains are of particular interest and were shown to have roles in signaling pathways and mediating network communication. The evolution of domain promiscuity is still an open problem, mostly due to the lack of sequenced ancestral genomes. Here we use inferred domain architectures of ancestral genomes to trace the evolution of domain promiscuity in eukaryotic genomes. We find an increase in average promiscuity along many branches of the eukaryotic tree. Moreover, domain promiscuity can proceed at almost a steady rate over long evolutionary time or exhibit lineage-specific acceleration. We also observe that many signaling and regulatory domains gained domain promiscuity around the Bilateria divergence. In addition we show that those domains that played a role in the creation of two body axes and existed before the divergence of the bilaterians from fungi/metazoan achieve a boost in their promiscuities during the bilaterian evolution.
Figures


References
-
- Koonin EV, Aravind L, Kondrashov AS. Cell. 2000;101:573–576. - PubMed
-
- Ekman D, Bjorklund AK, Frey-Skott J, Elofsson A. J Mol Biol. 2005;348:231–243. - PubMed
-
- Apic G, Gough J, Teichmann SA. J Mol Biol. 2001;310:311–325. - PubMed
-
- Apic G, Huber W, Teichmann SA. J Struct Funct Genomics. 2003;4:67–78. - PubMed
-
- Vogel C, Teichmann SA, Pereira-Leal J. J Mol Biol. 2005;346:355–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

