Studying the evolution of promoter sequences: a waiting time problem
- PMID: 21128851
- PMCID: PMC3119604
- DOI: 10.1089/cmb.2010.0084
Studying the evolution of promoter sequences: a waiting time problem
Abstract
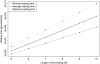
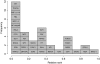
To gain a better understanding of the evolutionary dynamics of regulatory DNA sequences, we address the following questions: (1) How long does it take until a given transcription factor (TF) binding site emerges at random in a promoter sequence? and (2) How does the composition of a TF binding site affect this waiting time? Using two different probabilistic models (an i.i.d. model and a neighbor dependent model), we can compute the expected waiting time for every k-mer, k ranging from 5 to 10, until it appears in a promoter of a species. Our findings indicate that new TF binding sites can be created on a short evolutionary time scale, i.e. in a time span below the speciation time of human and chimp. Furthermore, one can conclude that the composition of a TF binding site plays a crucial role concerning the waiting time until it appears and that the CpG methylation-deamination substitution process probably accelerates the creation of new TF binding sites. A screening of existing TF binding sites moreover reveals that k-mers predicted to have short waiting times occur more frequently than others. Supplementary Material is available at www.libertonline.com/cmb .
Figures




References
-
- Arndt P.F. Burge C.B. Hwa T. DNA sequence evolution with neighbor-dependent mutation. J. Comput. Biol. 2003;10:313–322. - PubMed
-
- Arndt P.F. Hwa T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics. 2005;21:2322–2328. - PubMed
-
- Durrett R. Schmidt D. Waiting for regulatory sequences to appear. Annu. Appl. Probab. 2007;17:1–32.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
