The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations
- PMID: 21129061
- PMCID: PMC8094257
- DOI: 10.1111/j.1750-3639.2010.00454.x
The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations
Abstract
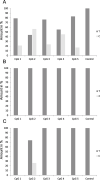
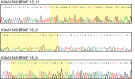
For some, glioma biomarkers have been expected to solve common diagnostic problems in routine neuropathology service caused by insufficient material, technical shortcomings or lack of experience. Further, biomarkers should predict patient outcome and direct optimal therapy for the individual patient. Unfortunately, current biomarkers still fall somewhat short of these grand expectations. While there has been some progress, it has generally been slow and in small steps. In this review, the newest set of glioma biomarkers: O(6) -methylguanine-DNA methyltransferase (MGMT) methylation, BRAF fusion and IDH1 mutation are discussed. MGMT methylation is well established as a prognostic/predictive marker for glioblastoma; however, technical questions regarding testing remain, it is not currently utilized widely in guiding patient management, and it has proven to be of no assistance in diagnostics. In contrast, BRAF fusion and IDH1 mutation analyses promise to be very helpful for classifying and grading gliomas, while their potential predictive value has yet to be established.
© 2010 The Authors; Brain Pathology © 2010 International Society of Neuropathology.
Figures


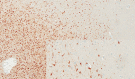
References
-
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, Von Deimling A (2008) Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116:597–602. - PubMed
-
- Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG (2008) Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol 67:878–887. - PubMed
-
- Belanich M, Pastor M, Randall T, Guerra D, Kibitel J, Alas L et al (1996) Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res 56:783–788. - PubMed
-
- Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP et al (2009) IDH1 mutations at residue p.R132 [IDH1(R132)] occur frequently in high‐grade gliomas but not in other solid tumors. Hum Mutat 30:7–11. - PubMed
-
- Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N et al (2010) Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol 28:3717–3723. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

