Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients
- PMID: 21129172
- PMCID: PMC3013085
- DOI: 10.1186/1471-2407-10-666
Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients
Abstract
Background: The presence of circulating tumor cells (CTC) in the peripheral blood of cancer patients has been described for various solid tumors and their clinical relevance has been shown. CTC detection based on the analysis of epithelial antigens might be hampered by the genetic heterogeneity of the primary tumor and loss of epithelial antigens. Therefore, we aimed to identify new gene markers for the PCR-based detection of CTC in female cancer patients.
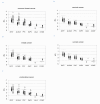
Methods: Gene expression of 38 cancer cell lines (breast, ovarian, cervical and endometrial) and of 10 peripheral blood mononuclear cell (PBMC) samples from healthy female donors was measured using microarray technology (Applied Biosystems). Differentially expressed genes were identified using the maxT test and the 50% one-sided trimmed maxT-test. Confirmatory RT-qPCR was performed for 380 gene targets using the AB TaqMan® Low Density Arrays. Then, 93 gene targets were analyzed using the same RT-qPCR platform in tumor tissues of 126 patients with primary breast, ovarian or endometrial cancer. Finally, blood samples from 26 healthy women and from 125 patients (primary breast, ovarian, cervical, or endometrial cancer, and advanced breast cancer) were analyzed following OncoQuick enrichment and RNA pre-amplification. Likewise, hMAM and EpCAM gene expression was analyzed in the blood of breast and ovarian cancer patients. For each gene, a cut-off threshold value was set at three standard deviations from the mean expression level of the healthy controls to identify potential markers for CTC detection.
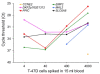
Results: Six genes were over-expressed in blood samples from 81% of patients with advanced and 29% of patients with primary breast cancer. EpCAM gene expression was detected in 19% and 5% of patients, respectively, whereas hMAM gene expression was observed in the advanced group (39%) only. Multimarker analysis using the new six gene panel positively identified 44% of the cervical, 64% of the endometrial and 19% of the ovarian cancer patients.
Conclusions: The panel of six genes was found superior to EpCAM and hMAM for the detection of circulating tumor cells in the blood of breast cancer, and they may serve as potential markers for CTC derived from endometrial, cervical, and ovarian cancers.
Figures

Similar articles
-
Circulating tumor cells (CTCs) detected by triple-marker EpCAM, CK19, and hMAM RT-PCR and their relation to clinical outcome in metastatic breast cancer patients.Cell Biochem Biophys. 2013 Mar;65(2):263-73. doi: 10.1007/s12013-012-9426-2. Cell Biochem Biophys. 2013. PMID: 22990361
-
Expression analysis of mammaglobin A (SCGB2A2) and lipophilin B (SCGB1D2) in more than 300 human tumors and matching normal tissues reveals their co-expression in gynecologic malignancies.BMC Cancer. 2006 Apr 9;6:88. doi: 10.1186/1471-2407-6-88. BMC Cancer. 2006. PMID: 16603086 Free PMC article.
-
EpCAM based capture detects and recovers circulating tumor cells from all subtypes of breast cancer except claudin-low.Oncotarget. 2015 Dec 29;6(42):44623-34. doi: 10.18632/oncotarget.5977. Oncotarget. 2015. PMID: 26556851 Free PMC article.
-
Real-time RT-PCR systems for CTC detection from blood samples of breast cancer and gynaecological tumour patients (Review).Oncol Rep. 2016 Apr;35(4):1905-15. doi: 10.3892/or.2016.4608. Epub 2016 Feb 2. Oncol Rep. 2016. PMID: 26848098 Review.
-
Current and future role of circulating tumor cells in patients with epithelial ovarian cancer.Eur J Surg Oncol. 2016 Dec;42(12):1772-1779. doi: 10.1016/j.ejso.2016.05.010. Epub 2016 May 25. Eur J Surg Oncol. 2016. PMID: 27265041 Review.
Cited by
-
Minimal residual disease in breast cancer: an overview of circulating and disseminated tumour cells.Clin Exp Metastasis. 2016 Aug;33(6):521-50. doi: 10.1007/s10585-016-9796-8. Epub 2016 May 17. Clin Exp Metastasis. 2016. PMID: 27189371 Free PMC article. Review.
-
Cancer Stem Cell-Like Circulating Tumor Cells Are Prognostic in Non-Small Cell Lung Cancer.J Pers Med. 2021 Nov 18;11(11):1225. doi: 10.3390/jpm11111225. J Pers Med. 2021. PMID: 34834576 Free PMC article.
-
Unlocking the Potential of Liquid Biopsy: A Paradigm Shift in Endometrial Cancer Care.Diagnostics (Basel). 2025 Jul 30;15(15):1916. doi: 10.3390/diagnostics15151916. Diagnostics (Basel). 2025. PMID: 40804879 Free PMC article. Review.
-
Identification of novel molecular markers for detection of gastric cancer cells in the peripheral blood circulation using genome-wide microarray analysis.Exp Ther Med. 2011 Jul;2(4):705-713. doi: 10.3892/etm.2011.252. Epub 2011 Apr 8. Exp Ther Med. 2011. PMID: 22977563 Free PMC article.
-
Circulating tumors cells as biomarkers: progress toward biomarker qualification.Cancer J. 2011 Nov-Dec;17(6):438-50. doi: 10.1097/PPO.0b013e31823e69ac. Cancer J. 2011. PMID: 22157288 Free PMC article. Review.
References
-
- GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. http://www-dep.iarc.fr
-
- Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aus Med J. 1869;14:146–149.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

