Structural analysis of bacteriophage T4 DNA replication: a review in the Virology Journal series on bacteriophage T4 and its relatives
- PMID: 21129204
- PMCID: PMC3012046
- DOI: 10.1186/1743-422X-7-359
Structural analysis of bacteriophage T4 DNA replication: a review in the Virology Journal series on bacteriophage T4 and its relatives
Abstract
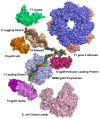
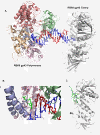
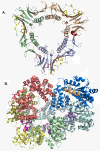
The bacteriophage T4 encodes 10 proteins, known collectively as the replisome, that are responsible for the replication of the phage genome. The replisomal proteins can be subdivided into three activities; the replicase, responsible for duplicating DNA, the primosomal proteins, responsible for unwinding and Okazaki fragment initiation, and the Okazaki repair proteins. The replicase includes the gp43 DNA polymerase, the gp45 processivity clamp, the gp44/62 clamp loader complex, and the gp32 single-stranded DNA binding protein. The primosomal proteins include the gp41 hexameric helicase, the gp61 primase, and the gp59 helicase loading protein. The RNaseH, a 5' to 3' exonuclease and T4 DNA ligase comprise the activities necessary for Okazaki repair. The T4 provides a model system for DNA replication. As a consequence, significant effort has been put forth to solve the crystallographic structures of these replisomal proteins. In this review, we discuss the structures that are available and provide comparison to related proteins when the T4 structures are unavailable. Three of the ten full-length T4 replisomal proteins have been determined; the gp59 helicase loading protein, the RNase H, and the gp45 processivity clamp. The core of T4 gp32 and two proteins from the T4 related phage RB69, the gp43 polymerase and the gp45 clamp are also solved. The T4 gp44/62 clamp loader has not been crystallized but a comparison to the E. coli gamma complex is provided. The structures of T4 gp41 helicase, gp61 primase, and T4 DNA ligase are unknown, structures from bacteriophage T7 proteins are discussed instead. To better understand the functionality of T4 DNA replication, in depth structural analysis will require complexes between proteins and DNA substrates. A DNA primer template bound by gp43 polymerase, a fork DNA substrate bound by RNase H, gp43 polymerase bound to gp32 protein, and RNase H bound to gp32 have been crystallographically determined. The preparation and crystallization of complexes is a significant challenge. We discuss alternate approaches, such as small angle X-ray and neutron scattering to generate molecular envelopes for modeling macromolecular assemblies.
Figures
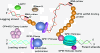


Similar articles
-
Coordinated DNA Replication by the Bacteriophage T4 Replisome.Viruses. 2015 Jun 19;7(6):3186-200. doi: 10.3390/v7062766. Viruses. 2015. PMID: 26102578 Free PMC article. Review.
-
Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome.Biochemistry. 2005 May 31;44(21):7747-56. doi: 10.1021/bi047296w. Biochemistry. 2005. PMID: 15909989
-
Control of helicase loading in the coupled DNA replication and recombination systems of bacteriophage T4.J Biol Chem. 2014 Jan 31;289(5):3040-54. doi: 10.1074/jbc.M113.505842. Epub 2013 Dec 14. J Biol Chem. 2014. PMID: 24338568 Free PMC article.
-
Investigation of stoichiometry of T4 bacteriophage helicase loader protein (gp59).J Biol Chem. 2009 Oct 23;284(43):29283-9. doi: 10.1074/jbc.M109.029926. Epub 2009 Aug 20. J Biol Chem. 2009. PMID: 19700405 Free PMC article.
-
DNA polymerase of the T4-related bacteriophages.Prog Nucleic Acid Res Mol Biol. 2000;64:65-96. doi: 10.1016/s0079-6603(00)64002-3. Prog Nucleic Acid Res Mol Biol. 2000. PMID: 10697407 Review.
Cited by
-
Assembly and subunit stoichiometry of the functional helicase-primase (primosome) complex of bacteriophage T4.Proc Natl Acad Sci U S A. 2012 Aug 21;109(34):13596-601. doi: 10.1073/pnas.1210040109. Epub 2012 Aug 6. Proc Natl Acad Sci U S A. 2012. PMID: 22869700 Free PMC article.
-
Clamp loader ATPases and the evolution of DNA replication machinery.BMC Biol. 2012 Apr 20;10:34. doi: 10.1186/1741-7007-10-34. BMC Biol. 2012. PMID: 22520345 Free PMC article.
-
Genome-wide characterization of Vibrio phage φpp2 with unique arrangements of the mob-like genes.BMC Genomics. 2012 Jun 7;13:224. doi: 10.1186/1471-2164-13-224. BMC Genomics. 2012. PMID: 22676552 Free PMC article.
-
Coordinated DNA Replication by the Bacteriophage T4 Replisome.Viruses. 2015 Jun 19;7(6):3186-200. doi: 10.3390/v7062766. Viruses. 2015. PMID: 26102578 Free PMC article. Review.
-
Gp2.5, the multifunctional bacteriophage T7 single-stranded DNA binding protein.Semin Cell Dev Biol. 2019 Feb;86:92-101. doi: 10.1016/j.semcdb.2018.03.018. Epub 2018 Mar 28. Semin Cell Dev Biol. 2019. PMID: 29588157 Free PMC article. Review.
References
-
- Watson JD, Crick FH. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953;18:123–131. - PubMed
-
- Kornberg A, Baker TA. DNA Replication. Second. New York: W.H. Freeman and Company; 1992.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

