Bayesian classification of residues associated with protein functional divergence: Arf and Arf-like GTPases
- PMID: 21129209
- PMCID: PMC3012027
- DOI: 10.1186/1745-6150-5-66
Bayesian classification of residues associated with protein functional divergence: Arf and Arf-like GTPases
Abstract
Background: Certain residues within proteins are highly conserved across very distantly related organisms, yet their (presumably critical) structural or mechanistic roles are completely unknown. To obtain clues regarding such residues within Arf and Arf-like (Arf/Arl) GTPases--which function as on/off switches regulating vesicle trafficking, phospholipid metabolism and cytoskeletal remodeling--I apply a new sampling procedure for comparative sequence analysis, termed multiple category Bayesian Partitioning with Pattern Selection (mcBPPS).
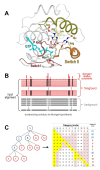
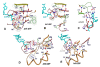
Results: The mcBPPS sampler classified sequences within the entire P-loop GTPase class into multiple categories by identifying those evolutionarily-divergent residues most likely to be responsible for functional specialization. Here I focus on categories of residues that most distinguish various Arf/Arl GTPases from other GTPases. This identified residues whose specific roles have been previously proposed (and in some cases corroborated experimentally and that thus serve as positive controls), as well as several categories of co-conserved residues whose possible roles are first hinted at here. For example, Arf/Arl/Sar GTPases are most distinguished from other GTPases by a conserved aspartate residue within the phosphate binding loop (P-loop) and by co-conserved residues nearby that, together, can form a network of salt-bridge and hydrogen bond interactions centered on the GTPase active site. Residues corresponding to an N-[VI] motif that is conserved within Arf/Arl GTPases may play a role in the interswitch toggle characteristic of the Arf family, whereas other, co-conserved residues may modulate the flexibility of the guanine binding loop. Arl8 GTPases conserve residues that strikingly diverge from those typically found in other Arf/Arl GTPases and that form structural interactions suggestive of a novel interswitch toggle mechanism.
Conclusions: This analysis suggests specific mutagenesis experiments to explore mechanisms underlying GTP hydrolysis, nucleotide exchange and interswitch toggling within Arf/Arl GTPases. More generally, it illustrates how the mcBPPS sampler can complement traditional evolutionary analyses by providing an objective, quantitative and statistically rigorous way to explore protein functional-divergence in molecular detail. Because the sampler classifies the input sequences at the same time, it can be used to generate subgroup profiles, in which functionally-divergent categories of residues are annotated automatically.
Figures



References
-
- Wittinghofer A. In: GTPases. Hall A, editor. Oxford: Oxford University Press; 2000. The Functioning of Molecular Switches in Three Dimensions; pp. 244–310.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

