An integrated approach to uncover drivers of cancer
- PMID: 21129771
- PMCID: PMC3013278
- DOI: 10.1016/j.cell.2010.11.013
An integrated approach to uncover drivers of cancer
Abstract
Systematic characterization of cancer genomes has revealed a staggering number of diverse aberrations that differ among individuals, such that the functional importance and physiological impact of most tumor genetic alterations remain poorly defined. We developed a computational framework that integrates chromosomal copy number and gene expression data for detecting aberrations that promote cancer progression. We demonstrate the utility of this framework using a melanoma data set. Our analysis correctly identified known drivers of melanoma and predicted multiple tumor dependencies. Two dependencies, TBC1D16 and RAB27A, confirmed empirically, suggest that abnormal regulation of protein trafficking contributes to proliferation in melanoma. Together, these results demonstrate the ability of integrative Bayesian approaches to identify candidate drivers with biological, and possibly therapeutic, importance in cancer.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
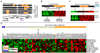
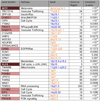

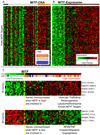
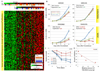
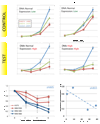
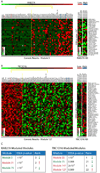
Comment in
-
Genomics: driving cancer biology.Nat Rev Cancer. 2011 Feb;11(2):79. Nat Rev Cancer. 2011. PMID: 21322290
References
-
- Beroukhim R, Brunet J-P, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. Patterns of Gene Expression and Copy-Number Alterations in von-Hippel Lindau Disease-Associated and Sporadic Clear Cell Carcinoma of the Kidney. Cancer Res. 2009;69:4674–4681. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

