Sequences at the interface of the fifth immunoglobulin domain and first fibronectin type III repeat of the neural cell adhesion molecule are critical for its polysialylation
- PMID: 21131353
- PMCID: PMC3039361
- DOI: 10.1074/jbc.M110.200386
Sequences at the interface of the fifth immunoglobulin domain and first fibronectin type III repeat of the neural cell adhesion molecule are critical for its polysialylation
Abstract
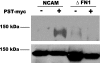
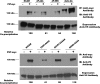
Polysialic acid is an anti-adhesive glycan that modifies a select group of mammalian proteins. The primary substrate of the polysialyltransferases (polySTs) is the neural cell adhesion molecule (NCAM). Polysialic acid negatively regulates cell adhesion, is required for proper brain development, and is expressed in specific areas of the adult brain where it promotes on-going cell migration and synaptic plasticity. The first fibronectin type III repeat (FN1) of NCAM is required for polysialylation of the N-glycans on the adjacent immunoglobulin-like domain (Ig5), and acidic residues on the surface of FN1 play a role in polyST recognition. Recent work demonstrated that the FN1 domain from the unpolysialylated olfactory cell adhesion molecule (OCAM) was able to partially replace NCAM FN1 (Foley, D. A., Swartzentruber, K. G., Thompson, M. G., Mendiratta, S. S., and Colley, K. J. (2010) J. Biol. Chem. 285, 35056-35067). Here we demonstrate that individually replacing three identical regions shared by NCAM and OCAM FN1, (500)PSSP(503) (PSSP), (526)GGVPI(530) (GGVPI), and (580)NGKG(583) (NGKG), dramatically reduces NCAM polysialylation. In addition, we show that the polyST, ST8SiaIV/PST, specifically binds NCAM and that this binding requires the FN1 domain. Replacing the FN1 PSSP sequences and the acidic patch residues decreases NCAM-polyST binding, whereas replacing the GGVPI and NGKG sequences has no effect. The location of GGVPI and NGKG in loops that flank the Ig5-FN1 linker and the proximity of PSSP to this linker suggest that GGVPI and NGKG sequences may be critical for stabilizing the Ig5-FN1 linker, whereas PSSP may play a dual role maintaining the Ig5-FN1 interface and a polyST recognition site.
Figures


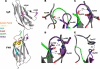



Similar articles
-
Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera.J Biol Chem. 2010 Nov 5;285(45):35056-67. doi: 10.1074/jbc.M110.170209. Epub 2010 Aug 30. J Biol Chem. 2010. PMID: 20805222 Free PMC article.
-
The polysialyltransferases interact with sequences in two domains of the neural cell adhesion molecule to allow its polysialylation.J Biol Chem. 2013 Mar 8;288(10):7282-93. doi: 10.1074/jbc.M112.438374. Epub 2013 Jan 22. J Biol Chem. 2013. PMID: 23341449 Free PMC article.
-
Specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule play a role in its recognition and polysialylation by the polysialyltransferase ST8Sia IV/PST.J Biol Chem. 2005 Sep 16;280(37):32340-8. doi: 10.1074/jbc.M506217200. Epub 2005 Jul 18. J Biol Chem. 2005. PMID: 16027151
-
A Possible Modulation Mechanism of Intramolecular and Intermolecular Interactions for NCAM Polysialylation and Cell Migration.Curr Top Med Chem. 2019;19(25):2271-2282. doi: 10.2174/1568026619666191018094805. Curr Top Med Chem. 2019. PMID: 31648641 Review.
-
3D structural conformation and functional domains of polysialyltransferase ST8Sia IV required for polysialylation of neural cell adhesion molecules.Protein Pept Lett. 2015;22(2):137-48. doi: 10.2174/0929866521666141019192221. Protein Pept Lett. 2015. PMID: 25329332 Review.
Cited by
-
Leucine-rich repeat neuronal protein 4 (LRRN4) potentially functions in dilated cardiomyopathy.Int J Clin Exp Pathol. 2017 Sep 1;10(9):9925-9933. eCollection 2017. Int J Clin Exp Pathol. 2017. PMID: 31966882 Free PMC article.
-
Glycobiology of neuroblastoma: impact on tumor behavior, prognosis, and therapeutic strategies.Front Oncol. 2014 May 23;4:114. doi: 10.3389/fonc.2014.00114. eCollection 2014. Front Oncol. 2014. PMID: 24904828 Free PMC article. Review.
-
Is Polysialylated NCAM Not Only a Regulator during Brain Development But also during the Formation of Other Organs?Biology (Basel). 2017 Apr 27;6(2):27. doi: 10.3390/biology6020027. Biology (Basel). 2017. PMID: 28448440 Free PMC article. Review.
-
Sequences prior to conserved catalytic motifs of polysialyltransferase ST8Sia IV are required for substrate recognition.J Biol Chem. 2012 Feb 24;287(9):6441-53. doi: 10.1074/jbc.M111.322024. Epub 2011 Dec 19. J Biol Chem. 2012. PMID: 22184126 Free PMC article.
-
The Polybasic Region of the Polysialyltransferase ST8Sia-IV Binds Directly to the Neural Cell Adhesion Molecule, NCAM.Biochemistry. 2017 Mar 14;56(10):1504-1517. doi: 10.1021/acs.biochem.6b01221. Epub 2017 Mar 3. Biochemistry. 2017. PMID: 28233978 Free PMC article.
References
-
- Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. (1987) Science 236, 799–806 - PubMed
-
- Walmod P. S., Kolkova K., Berezin V., Bock E. (2004) Neurochem. Res. 29, 2015–2035 - PubMed
-
- Ditlevsen D. K., Povlsen G. K., Berezin V., Bock E. (2008) J. Neurosci. Res. 86, 727–743 - PubMed
-
- Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Nature 373, 715–718 - PubMed
-
- Kojima N., Yoshida Y., Tsuji S. (1995) FEBS Lett. 373, 119–122 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

