The role of germline promoters and I exons in cytokine-induced gene-specific class switch recombination
- PMID: 21131417
- PMCID: PMC3059604
- DOI: 10.4049/jimmunol.1003108
The role of germline promoters and I exons in cytokine-induced gene-specific class switch recombination
Abstract
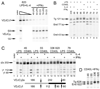
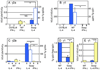
Germline transcription precedes class switch recombination (CSR). The promoter regions and I exons of these germline transcripts include binding sites for activation- and cytokine-induced transcription factors, and the promoter regions/I exons are essential for CSR. Therefore, it is a strong hypothesis that the promoter/I exons regions are responsible for much of cytokine-regulated, gene-specific CSR. We tested this hypothesis by swapping the germline promoter and I exons for the murine γ1 and γ2a H chain genes in a transgene of the entire H chain C-region locus. We found that the promoter/I exon for γ1 germline transcripts can direct robust IL-4-induced recombination to the γ2a gene. In contrast, the promoter/I exon for the γ2a germline transcripts works poorly in the context of the γ1 H chain gene, resulting in expression of γ1 H chains that is <1% the wild-type level. Nevertheless, the small amount of recombination to the chimeric γ1 gene is induced by IFN-γ. These results suggest that cytokine regulation of CSR, but not the magnitude of CSR, is regulated by the promoter/I exons.
Figures




References
-
- Stavnezer J. Molecular processes that regulate class-switching. Curr.Topics Micro.Immunol. 2000;245:127–168. - PubMed
-
- Cogne M, Birshtein BK. Regulation of class switch recombination. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. London: Academic Press; 2004. pp. 289–305.
-
- Bergstedt-Lindqvist S, Sideras P, MacDonald HR, Severinson E. Regulation of Ig class secretion by soluble products of certain T-cell lines. Immun.Rev. 1989;78:25–50. - PubMed
-
- Snapper CM, Paul WE. Interferon-γ and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

