Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata
- PMID: 21131438
- PMCID: PMC3067410
- DOI: 10.1128/EC.00277-10
Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata
Abstract
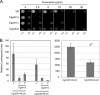
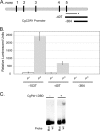
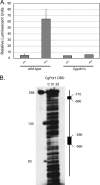
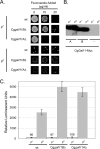
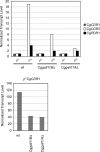
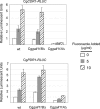
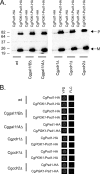
Candida glabrata is an opportunistic human pathogen that is increasingly associated with candidemia, owing in part to the intrinsic and acquired high tolerance the organism exhibits for the important clinical antifungal drug fluconazole. This elevated fluconazole resistance often develops through gain-of-function mutations in the zinc cluster-containing transcriptional regulator C. glabrata Pdr1 (CgPdr1). CgPdr1 induces the expression of an ATP-binding cassette (ABC) transporter-encoding gene, CgCDR1. Saccharomyces cerevisiae has two CgPdr1 homologues called ScPdr1 and ScPdr3. These factors control the expression of an ABC transporter-encoding gene called ScPDR5, which encodes a homologue of CgCDR1. Loss of the mitochondrial genome (ρ(0) cell) or overexpression of the mitochondrial enzyme ScPsd1 induces ScPDR5 expression in a strictly ScPdr3-dependent fashion. ScPdr3 requires the presence of a transcriptional Mediator subunit called Gal11 (Med15) to fully induce ScPDR5 transcription in response to ρ(0) signaling. ScPdr1 does not respond to either ρ(0) signals or ScPsd1 overproduction. In this study, we employed transcriptional fusions between CgPdr1 target promoters, like CgCDR1, to demonstrate that CgPdr1 stimulates gene expression via binding to elements called pleiotropic drug response elements (PDREs). Deletion mapping and electrophoretic mobility shift assays demonstrated that a single PDRE in the CgCDR1 promoter was capable of supporting ρ(0)-induced gene expression. Removal of one of the two ScGal11 homologues from C. glabrata caused a major defect in drug-induced expression of CgCDR1 but had a quantitatively minor effect on ρ(0)-stimulated transcription. These data demonstrate that CgPdr1 appears to combine features of ScPdr1 and ScPdr3 to produce a transcription factor with chimeric regulatory properties.
Figures
Similar articles
-
Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata.PLoS One. 2011 Mar 9;6(3):e17589. doi: 10.1371/journal.pone.0017589. PLoS One. 2011. PMID: 21408004 Free PMC article.
-
Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants.Antimicrob Agents Chemother. 2006 Apr;50(4):1384-92. doi: 10.1128/AAC.50.4.1384-1392.2006. Antimicrob Agents Chemother. 2006. PMID: 16569856 Free PMC article.
-
Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence.PLoS Pathog. 2009 Jan;5(1):e1000268. doi: 10.1371/journal.ppat.1000268. Epub 2009 Jan 16. PLoS Pathog. 2009. PMID: 19148266 Free PMC article.
-
Multiple interfaces control activity of the Candida glabrata Pdr1 transcription factor mediating azole drug resistance.Curr Genet. 2019 Feb;65(1):103-108. doi: 10.1007/s00294-018-0870-4. Epub 2018 Jul 28. Curr Genet. 2019. PMID: 30056490 Free PMC article. Review.
-
From Saccharomyces cerevisiae to Candida glabratain a few easy steps: important adaptations for an opportunistic pathogen.FEMS Microbiol Lett. 2011 Jan;314(1):1-9. doi: 10.1111/j.1574-6968.2010.02102.x. Epub 2010 Sep 16. FEMS Microbiol Lett. 2011. PMID: 20846362 Free PMC article. Review.
Cited by
-
Jjj1 Is a Negative Regulator of Pdr1-Mediated Fluconazole Resistance in Candida glabrata.mSphere. 2018 Feb 21;3(1):e00466-17. doi: 10.1128/mSphere.00466-17. eCollection 2018 Jan-Feb. mSphere. 2018. PMID: 29507891 Free PMC article.
-
Candida glabrata Biofilms: How Far Have We Come?J Fungi (Basel). 2017 Mar 1;3(1):11. doi: 10.3390/jof3010011. J Fungi (Basel). 2017. PMID: 29371530 Free PMC article. Review.
-
Genetic interaction analysis of Candida glabrata transcription factors CST6 and UPC2A in the regulation of respiration and fluconazole susceptibility.Antimicrob Agents Chemother. 2025 Feb 13;69(2):e0129424. doi: 10.1128/aac.01294-24. Epub 2024 Dec 23. Antimicrob Agents Chemother. 2025. PMID: 39714155 Free PMC article.
-
Identification and Characterization of Key Charged Residues in the Cofilin Protein Involved in Azole Susceptibility, Apoptosis, and Virulence of Aspergillus fumigatus.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e01659-17. doi: 10.1128/AAC.01659-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29483117 Free PMC article.
-
Update on Antifungal Drug Resistance.Curr Clin Microbiol Rep. 2015 Jun 1;2(2):84-95. doi: 10.1007/s40588-015-0015-1. Curr Clin Microbiol Rep. 2015. PMID: 26120512 Free PMC article.
References
-
- Balzi E., Goffeau A. 1991. Multiple or pleiotropic drug resistance in yeast. BBA 1073:241–252 - PubMed
-
- Balzi E., Wang M., Leterme S., Van Dyck L., Goffeau A. 1994. PDR5: a novel yeast multidrug resistance transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269:2206–2214 - PubMed
-
- Bissinger P. H., Kuchler K. 1994. Molecular cloning and expression of the S. cerevisiae STS1 gene product. J. Biol. Chem. 269:4180–4186 - PubMed
-
- Bourbon H. M., et al. 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14:553–557 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

