Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence
- PMID: 21131439
- PMCID: PMC3067405
- DOI: 10.1128/EC.00158-10
Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence
Abstract
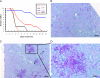
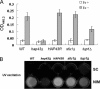
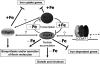
Candida albicans is an opportunistic fungal pathogen that exists as normal flora in healthy human bodies but causes life-threatening infections in immunocompromised patients. In addition to innate and adaptive immunities, hosts also resist microbial infections by developing a mechanism of "natural resistance" that maintains a low level of free iron to restrict the growth of invading pathogens. C. albicans must overcome this iron-deprived environment to cause infections. There are three types of iron-responsive transcriptional regulators in fungi; Aft1/Aft2 activators in yeast, GATA-type repressors in many fungi, and HapX/Php4 in Schizosaccharomyces pombe and Aspergillus species. In this study, we characterized the iron-responsive regulator Hap43, which is the C. albicans homolog of HapX/Php4 and is repressed by the GATA-type repressor Sfu1 under iron-sufficient conditions. We provide evidence that Hap43 is essential for the growth of C. albicans under low-iron conditions and for C. albicans virulence in a mouse model of infection. Hap43 was not required for iron acquisition under low-iron conditions. Instead, it was responsible for repression of genes that encode iron-dependent proteins involved in mitochondrial respiration and iron-sulfur cluster assembly. We also demonstrated that Hap43 executes its function by becoming a transcriptional repressor and accumulating in the nucleus in response to iron deprivation. Finally, we found a connection between Hap43 and the global corepressor Tup1 in low-iron-induced flavinogenesis. Taken together, our data suggest a complex interplay among Hap43, Sfu1, and Tup1 to coordinately regulate iron acquisition, iron utilization, and other iron-responsive metabolic activities.
Figures





References
-
- Archibald F. 1983. Lactobacillus plantarum, an organism not requiring iron. FEMS Microbiol. Lett. 19:29–32
-
- Blaiseau P. L., Lesuisse E., Camadro J. M. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276:34221–34226 - PubMed
-
- Boretsky Y. R., et al. 2005. Positive selection of mutants defective in transcriptional repression of riboflavin synthesis by iron in the flavinogenic yeast Pichia guilliermondii. FEMS Yeast Res. 5:829–837 - PubMed
-
- Bourgarel D., Nguyen C. C., Bolotin-Fukuhara M. 1999. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 31:1205–1215 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

