A single cell functions as a tissue-specific stem cell and the in vitro niche-forming cell
- PMID: 21131442
- PMCID: PMC3175586
- DOI: 10.1165/rcmb.2010-0314OC
A single cell functions as a tissue-specific stem cell and the in vitro niche-forming cell
Abstract
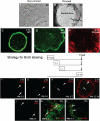
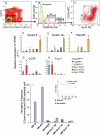
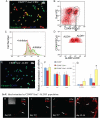
Tissue-specific stem cell (TSC) behavior is determined by the stem cell niche. However, delineation of the TSC-niche interaction requires purification of both entities. We reasoned that the niche could be defined by the location of the TSC. We demonstrate that a single CD49f(bright)/Sca1(+)/ALDH(+) basal cell generates rare label-retaining cells and abundant label-diluting cells. Label-retaining and label-diluting cells were located in the rimmed domain of a unique clone type, the rimmed clone. The TSC property of self-renewal was tested by serial passage at clonal density and analysis of clone-forming cell frequency. A single clone could be passaged up to five times and formed only rimmed clones. Thus, rimmed clone formation was a cell-intrinsic property. Differentiation potential was evaluated in air-liquid interface cultures. Homogenous cultures of rimmed clones were highly mitotic but were refractory to standard differentiation signals. However, rimmed clones that were cocultured with unfractionated tracheal cells generated each of the cell types found in the tracheal epithelium. Thus, the default niche is promitotic: Multipotential differentiation requires adaptation of the niche. Because lung TSCs are typically evaluated after injury, the behavior of CD49f(bright)/Sca1(+)/ALDH(+) cells was tested in normal and naphthalene-treated mice. These cells were mitotically active in the normal and repaired epithelium, their proliferation rate increased in response to injury, and they retained label for 34 days. We conclude that the CD49f(bright)/Sca1(+)/ALDH(+) tracheal basal cell is a TSC, that it generates its own niche in vitro, and that it participates in tracheal epithelial homeostasis and repair.
Figures


References
-
- Morrison SJ. Stem cell potential: can anything make anything?. Curr Biol 2001;11:R7–R9 - PubMed
-
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978;4:7–25 - PubMed
-
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 2004;118:635–648 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

