A caffeyl-coenzyme A synthetase initiates caffeate activation prior to caffeate reduction in the acetogenic bacterium Acetobacterium woodii
- PMID: 21131487
- PMCID: PMC3028691
- DOI: 10.1128/JB.01126-10
A caffeyl-coenzyme A synthetase initiates caffeate activation prior to caffeate reduction in the acetogenic bacterium Acetobacterium woodii
Abstract
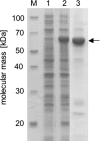
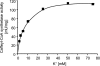
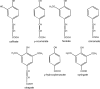
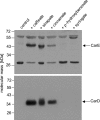
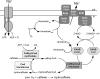
The anaerobic acetogenic bacterium Acetobacterium woodii couples the reduction of caffeate with electrons derived from hydrogen to the synthesis of ATP by a chemiosmotic mechanism using sodium ions as coupling ions, but the enzymes involved remain to be established. Previously, the electron transfer flavoproteins EtfA and EtfB were found to be involved in caffeate respiration. By inverse PCR, we identified three genes upstream of etfA and etfB: carA, carB, and carC. carA encodes a potential coenzyme A (CoA) transferase, carB an acyl-CoA synthetase, and carC an acyl-CoA dehydrogenase. carA, -B, and -C are located together with etfA/carE and etfB/carD on one polycistronic message, indicating that CarA, CarB, and CarC are also part of the caffeate respiration pathway. The genetic data suggest an initial ATP-dependent activation of caffeate by CarB. To prove the proposed function of CarB, the protein was overproduced in Escherichia coli, and the recombinant protein was purified. Purified CarB activates caffeate to caffeyl-CoA in an ATP- and CoA-dependent reaction. The enzyme has broad pH and temperature optima and requires K(+) for activity. In addition to caffeate, it can use ρ-coumarate, ferulate, and cinnamate as substrates, with 50, 15, and 9%, respectively, of the activity obtained with caffeate. Expression of the car operon is induced not only by caffeate, ρ-coumarate, ferulate, and cinnamate but also by sinapate. There is no induction by ρ-hydroxybenzoate or syringate.
Figures



References
-
- Aufurth, S., H. Schägger, and V. Müller. 2000. Identification of subunits a, b, and c1 from Acetobacterium woodii Na+-F1FO-ATPase. Subunits c1, c2, and c3 constitute a mixed c-oligomer. J. Biol. Chem. 275:33297-33301. - PubMed
-
- Bache, R., and N. Pfennig. 1981. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch. Microbiol. 130:255-261.
-
- Beaty, P. S., and L. G. Ljungdahl. 1991. Growth of Clostridium thermoaceticum on methanol, ethanol or dimethylsulfoxide, abstr. K-131, p. 236. Abstr. 91st Annu. Meet. Am. Soc. Microbiol. 1991. American Society for Microbiology, Washington, DC.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

