DifA, a methyl-accepting chemoreceptor protein-like sensory protein, uses a novel signaling mechanism to regulate exopolysaccharide production in Myxococcus xanthus
- PMID: 21131490
- PMCID: PMC3021235
- DOI: 10.1128/JB.00944-10
DifA, a methyl-accepting chemoreceptor protein-like sensory protein, uses a novel signaling mechanism to regulate exopolysaccharide production in Myxococcus xanthus
Abstract
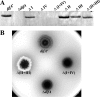
DifA is a methyl-accepting chemotaxis protein (MCP)-like sensory transducer that regulates exopolysaccharide (EPS) production in Myxococcus xanthus. Here mutational analysis and molecular biology were used to probe the signaling mechanisms of DifA in EPS regulation. We first identified the start codon of DifA experimentally; this identification extended the N terminus of DifA for 45 amino acids (aa) from the previous bioinformatics prediction. This extension helped to address the outstanding question of how DifA receives input signals from type 4 pili without a prominent periplasmic domain. The results suggest that DifA uses its N-terminus extension to sense an upstream signal in EPS regulation. We suggest that the perception of the input signal by DifA is mediated by protein-protein interactions with upstream components. Subsequent signal transmission likely involves transmembrane signaling instead of direct intramolecular interactions between the input and the output modules in the cytoplasm. The basic functional unit of DifA for signal transduction is likely dimeric as mutational alteration of the predicted dimeric interface of DifA significantly affected EPS production. Deletions of 14-aa segments in the C terminus suggest that the newly defined flexible bundle subdomain in MCPs is likely critical for DifA function because shortening of this bundle can lead to constitutively active mutations.
Figures


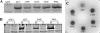


Similar articles
-
Isolation and characterization of a suppressor mutation that restores Myxococcus xanthus exopolysaccharide production.Microbiology (Reading). 2009 Nov;155(Pt 11):3599-3610. doi: 10.1099/mic.0.031070-0. Epub 2009 Aug 14. Microbiology (Reading). 2009. PMID: 19684067 Free PMC article.
-
Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exopolysaccharide regulation in Myxococcus xanthus.J Bacteriol. 2010 Sep;192(17):4267-74. doi: 10.1128/JB.00403-10. Epub 2010 Jun 11. J Bacteriol. 2010. PMID: 20543066 Free PMC article.
-
Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX-DifA interspecies chimera.J Bacteriol. 2005 Sep;187(18):6410-8. doi: 10.1128/JB.187.18.6410-6418.2005. J Bacteriol. 2005. PMID: 16159775 Free PMC article.
-
Phospholipid directed motility of surface-motile bacteria.Mol Microbiol. 2006 Sep;61(5):1101-9. doi: 10.1111/j.1365-2958.2006.05314.x. Mol Microbiol. 2006. PMID: 16925549 Review.
-
Dual regulation with Ser/Thr kinase cascade and a His/Asp TCS in Myxococcus xanthus.Adv Exp Med Biol. 2008;631:111-21. doi: 10.1007/978-0-387-78885-2_7. Adv Exp Med Biol. 2008. PMID: 18792684 Review.
Cited by
-
Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility.Proc Natl Acad Sci U S A. 2012 Jun 19;109(25):10036-41. doi: 10.1073/pnas.1120979109. Epub 2012 Jun 4. Proc Natl Acad Sci U S A. 2012. PMID: 22665761 Free PMC article.
-
MasABK proteins interact with proteins of the type IV pilin system to affect social motility of Myxococcus xanthus.PLoS One. 2013;8(1):e54557. doi: 10.1371/journal.pone.0054557. Epub 2013 Jan 16. PLoS One. 2013. PMID: 23342171 Free PMC article.
-
Mediation of Extracellular Polymeric Substances in Microbial Reduction of Hematite by Shewanella oneidensis MR-1.Front Microbiol. 2019 Mar 29;10:575. doi: 10.3389/fmicb.2019.00575. eCollection 2019. Front Microbiol. 2019. PMID: 30984128 Free PMC article.
References
-
- Alexander, R. 2007. Evolutionary genomics of methyl-accepting chemotaxis proteins. Georgia Institute of Technology, Atlanta, GA.
-
- Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. - PubMed
-
- Ayers, M., P. L. Howell, and L. L. Burrows. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5:1203-1218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

