Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase
- PMID: 21131906
- PMCID: PMC3025462
- DOI: 10.1038/emboj.2010.317
Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase
Abstract
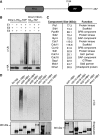
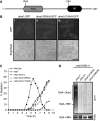
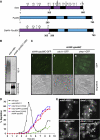
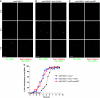
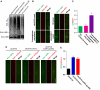
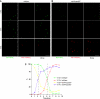
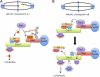
Proper cell division requires strict coordination between mitotic exit and cytokinesis. In the event of a mitotic error, cytokinesis must be inhibited to ensure equal partitioning of genetic material. In the fission yeast, Schizosaccharomyces pombe, the checkpoint protein and E3 ubiquitin ligase, Dma1, delays cytokinesis by inhibiting the septation initiation network (SIN) when chromosomes are not attached to the mitotic spindle. To elucidate the mechanism by which Dma1 inhibits the SIN, we screened all SIN components as potential Dma1 substrates and found that the SIN scaffold protein, Sid4, is ubiquitinated in vivo in a Dma1-dependent manner. To investigate the role of Sid4 ubiquitination in checkpoint function, a ubiquitination deficient sid4 allele was generated and our data indicate that Sid4 ubiquitination by Dma1 is required to prevent cytokinesis during a mitotic checkpoint arrest. Furthermore, Sid4 ubiquitination delays recruitment of the Polo-like kinase and SIN activator, Plo1, to spindle pole bodies (SPBs), while at the same time prolonging residence of the SIN inhibitor, Byr4, providing a mechanistic link between Dma1 activity and cytokinesis inhibition.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
CK1 is required for a mitotic checkpoint that delays cytokinesis.Curr Biol. 2013 Oct 7;23(19):1920-6. doi: 10.1016/j.cub.2013.07.077. Epub 2013 Sep 19. Curr Biol. 2013. PMID: 24055157 Free PMC article.
-
Relief of the Dma1-mediated checkpoint requires Dma1 autoubiquitination and dynamic localization.Mol Biol Cell. 2018 Sep 1;29(18):2176-2189. doi: 10.1091/mbc.E18-04-0261. Epub 2018 Jul 5. Mol Biol Cell. 2018. PMID: 29975113 Free PMC article.
-
Dnt1 acts as a mitotic inhibitor of the spindle checkpoint protein dma1 in fission yeast.Mol Biol Cell. 2012 Sep;23(17):3348-56. doi: 10.1091/mbc.E11-12-1020. Epub 2012 Jul 18. Mol Biol Cell. 2012. PMID: 22809626 Free PMC article.
-
Spatiotemporal regulation of the Dma1-mediated mitotic checkpoint coordinates mitosis with cytokinesis.Curr Genet. 2019 Jun;65(3):663-668. doi: 10.1007/s00294-018-0921-x. Epub 2019 Jan 2. Curr Genet. 2019. PMID: 30600396 Free PMC article. Review.
-
Polar opposites: Fine-tuning cytokinesis through SIN asymmetry.Cytoskeleton (Hoboken). 2012 Oct;69(10):686-99. doi: 10.1002/cm.21044. Epub 2012 Jul 11. Cytoskeleton (Hoboken). 2012. PMID: 22786806 Free PMC article. Review.
Cited by
-
The kinase domain of CK1 enzymes contains the localization cue essential for compartmentalized signaling at the spindle pole.Mol Biol Cell. 2018 Jul 1;29(13):1664-1674. doi: 10.1091/mbc.E18-02-0129. Epub 2018 May 9. Mol Biol Cell. 2018. PMID: 29742018 Free PMC article.
-
Dma/RNF8 proteins are evolutionarily conserved E3 ubiquitin ligases that target septins.Cell Cycle. 2013 Mar 15;12(6):1000-8. doi: 10.4161/cc.23947. Epub 2013 Feb 26. Cell Cycle. 2013. PMID: 23442799 Free PMC article.
-
Fission yeast nucleolar protein Dnt1 regulates G2/M transition and cytokinesis by downregulating Wee1 kinase.J Cell Sci. 2013 Nov 1;126(Pt 21):4995-5004. doi: 10.1242/jcs.132845. Epub 2013 Sep 4. J Cell Sci. 2013. PMID: 24006256 Free PMC article.
-
CK1 is required for a mitotic checkpoint that delays cytokinesis.Curr Biol. 2013 Oct 7;23(19):1920-6. doi: 10.1016/j.cub.2013.07.077. Epub 2013 Sep 19. Curr Biol. 2013. PMID: 24055157 Free PMC article.
-
Localization of the ubiquitin ligase Dma1 to the fission yeast contractile ring is modulated by phosphorylation.FEBS Lett. 2021 Nov;595(22):2781-2792. doi: 10.1002/1873-3468.14211. Epub 2021 Nov 2. FEBS Lett. 2021. PMID: 34674264 Free PMC article.
References
-
- Bardin AJ, Amon A (2001) Men and sin: what's the difference? Nat Rev Mol Cell Biol 2: 815–826 - PubMed
-
- Beltraminelli N, Murone M, Simanis V (1999) The S. pombe zfs1 gene is required to prevent septation if mitotic progression is inhibited. J Cell Sci 112(Pt 18): 3103–3114 - PubMed
-
- Bothos J, Summers MK, Venere M, Scolnick DM, Halazonetis TD (2003) The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene 22: 7101–7107 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

