HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide
- PMID: 21131964
- PMCID: PMC3018327
- DOI: 10.1038/ni.1967
HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide
Abstract
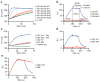
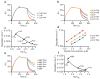
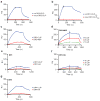
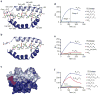
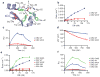
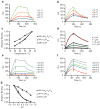
The mechanisms of HLA-DM-catalyzed peptide exchange remain uncertain. Here we found that all stages of the interaction of HLA-DM with HLA-DR were dependent on the occupancy state of the peptide-binding groove. High-affinity peptides were protected from removal by HLA-DM through two mechanisms: peptide binding induced the dissociation of a long-lived complex of empty HLA-DR and HLA-DM, and high-affinity HLA-DR-peptide complexes bound HLA-DM only very slowly. Nonbinding covalent HLA-DR-peptide complexes were converted into efficient HLA-DM binders after truncation of an N-terminal peptide segment that emptied the P1 pocket and disrupted conserved hydrogen bonds to HLA-DR. HLA-DM thus binds only to HLA-DR conformers in which a critical part of the binding site is already vacant because of spontaneous peptide motion.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
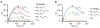
References
-
- Lanzavecchia A, Reid PA, Watts C. Irreversible association of peptides with class II MHC molecules in living cells. Nature. 1992;357:249–252. - PubMed
-
- Stern LJ, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. - PubMed
-
- Brown JH, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. - PubMed
-
- Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

