An assessment of histone-modification antibody quality
- PMID: 21131980
- PMCID: PMC3017233
- DOI: 10.1038/nsmb.1972
An assessment of histone-modification antibody quality
Abstract
We have tested the specificity and utility of more than 200 antibodies raised against 57 different histone modifications in Drosophila melanogaster, Caenorhabditis elegans and human cells. Although most antibodies performed well, more than 25% failed specificity tests by dot blot or western blot. Among specific antibodies, more than 20% failed in chromatin immunoprecipitation experiments. We advise rigorous testing of histone-modification antibodies before use, and we provide a website for posting new test results (http://compbio.med.harvard.edu/antibodies/).
Conflict of interest statement
Fifteen non-commercial antibodies (indicated in Supplementary Table 1) were provided to us at no cost. We have no financial or material interest in the companies whose antibodies were tested.
Figures
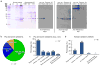


References
-
- Sourkes TL. 1901–1921. Nobel Lectures. Including presentation speeches and laureates’ biographies. Elsevier Publishing Co; Amsterdam, London, and New York: 1967.
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

