The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules
- PMID: 21131981
- PMCID: PMC3056208
- DOI: 10.1038/nsmb.1936
The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules
Abstract
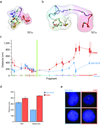
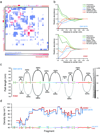
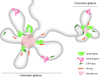
We developed a general approach that combines chromosome conformation capture carbon copy (5C) with the Integrated Modeling Platform (IMP) to generate high-resolution three-dimensional models of chromatin at the megabase scale. We applied this approach to the ENm008 domain on human chromosome 16, containing the α-globin locus, which is expressed in K562 cells and silenced in lymphoblastoid cells (GM12878). The models accurately reproduce the known looping interactions between the α-globin genes and their distal regulatory elements. Further, we find using our approach that the domain folds into a single globular conformation in GM12878 cells, whereas two globules are formed in K562 cells. The central cores of these globules are enriched for transcribed genes, whereas nontranscribed chromatin is more peripheral. We propose that globule formation represents a higher-order folding state related to clustering of transcribed genes around shared transcription machineries, as previously observed by microscopy.
Figures



References
-
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. - PubMed
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. - PubMed
-
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

