CtIP promotes microhomology-mediated alternative end joining during class-switch recombination
- PMID: 21131982
- PMCID: PMC3471154
- DOI: 10.1038/nsmb.1942
CtIP promotes microhomology-mediated alternative end joining during class-switch recombination
Abstract
Immunoglobulin heavy chain (Igh locus) class-switch recombination (CSR) requires targeted introduction of DNA double strand breaks (DSBs) into repetitive 'switch'-region DNA elements in the Igh locus and subsequent ligation between distal DSBs. Both canonical nonhomologous end joining (C-NHEJ) that seals DNA ends with little or no homology and a poorly defined alternative end joining (A-NHEJ, also known as alt-NHEJ) process that requires microhomology ends for ligation have been implicated in CSR. Here, we show that the DNA end-processing factor CtIP is required for microhomology-directed A-NHEJ during CSR. Additionally, we demonstrate that microhomology joins that are enriched upon depletion of the C-NHEJ component Ku70 require CtIP. Finally, we show that CtIP binds to switch-region DNA in a fashion dependent on activation-induced cytidine deaminase. Our results establish CtIP as a bona fide component of microhomology-dependent A-NHEJ and unmask a hitherto unrecognized physiological role of microhomology-mediated end joining in a C-NHEJ-proficient environment.
Figures
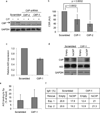

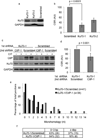


Similar articles
-
IgH class switching and translocations use a robust non-classical end-joining pathway.Nature. 2007 Sep 27;449(7161):478-82. doi: 10.1038/nature06020. Epub 2007 Aug 22. Nature. 2007. PMID: 17713479
-
CtIP-mediated DNA resection is dispensable for IgH class switch recombination by alternative end-joining.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25700-25711. doi: 10.1073/pnas.2010972117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989150 Free PMC article.
-
Alternative end-joining and classical nonhomologous end-joining pathways repair different types of double-strand breaks during class-switch recombination.J Immunol. 2013 Dec 1;191(11):5751-63. doi: 10.4049/jimmunol.1301300. Epub 2013 Oct 21. J Immunol. 2013. PMID: 24146042
-
Non-homologous end joining in class switch recombination: the beginning of the end.Philos Trans R Soc Lond B Biol Sci. 2009 Mar 12;364(1517):653-65. doi: 10.1098/rstb.2008.0196. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19008195 Free PMC article. Review.
-
Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks.Adv Immunol. 2012;116:1-49. doi: 10.1016/B978-0-12-394300-2.00001-6. Adv Immunol. 2012. PMID: 23063072 Review.
Cited by
-
DNA double-strand breaks and telomeres play important roles in trypanosoma brucei antigenic variation.Eukaryot Cell. 2015 Mar;14(3):196-205. doi: 10.1128/EC.00207-14. Epub 2015 Jan 9. Eukaryot Cell. 2015. PMID: 25576484 Free PMC article. Review.
-
Role of EXO1 nuclease activity in genome maintenance, the immune response and tumor suppression in Exo1D173A mice.Nucleic Acids Res. 2022 Aug 12;50(14):8093-8106. doi: 10.1093/nar/gkac616. Nucleic Acids Res. 2022. PMID: 35849338 Free PMC article.
-
Combinatorial mechanisms regulating AID-dependent DNA deamination: interacting proteins and post-translational modifications.Semin Immunol. 2012 Aug;24(4):264-72. doi: 10.1016/j.smim.2012.05.006. Epub 2012 Jul 6. Semin Immunol. 2012. PMID: 22771392 Free PMC article. Review.
-
Generation and repair of AID-initiated DNA lesions in B lymphocytes.Front Med. 2014 Jun;8(2):201-16. doi: 10.1007/s11684-014-0324-4. Epub 2014 Apr 21. Front Med. 2014. PMID: 24748462 Free PMC article. Review.
-
Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining.Am J Cancer Res. 2012;2(3):249-68. Epub 2012 Apr 21. Am J Cancer Res. 2012. PMID: 22679557 Free PMC article.
References
-
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. - PubMed
-
- Zarrin AA, et al. Antibody class switching mediated by yeast endonuclease generated DNA breaks. Science. 2007;315:377–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

