Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi
- PMID: 21131983
- PMCID: PMC3394154
- DOI: 10.1038/nbt.1720
Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi
Abstract
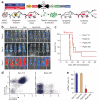
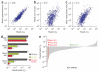
Short hairpin RNAs (shRNAs) are versatile tools for analyzing loss-of-function phenotypes in vitro and in vivo. However, their use for studying genes involved in proliferation and survival, which are potential therapeutic targets in cancer and other diseases, is confounded by the strong selective advantage of cells in which shRNA expression is inefficient. We therefore developed a toolkit that combines Tet-regulated miR30-shRNA technology, robust transactivator expression and two fluorescent reporters to track and isolate cells with potent target knockdown. We demonstrated that this system improves the study of essential genes and was sufficiently robust to eradicate aggressive cancer in mice by suppressing a single gene. Further, we applied this system for in vivo negative-selection screening with pooled shRNAs and propose a streamlined, inexpensive workflow that will facilitate the use of RNA interference (RNAi) for the identification and evaluation of essential therapeutic targets.
Figures


References
-
- Huesken D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. - PubMed
-
- McCurrach ME, Lowe SW. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 2001;66:197–227. - PubMed
-
- Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

