GOAPhAR: An Integrative Discovery Tool for Annotation, Pathway Analysis
- PMID: 21132056
- PMCID: PMC2995274
- DOI: 10.2174/1875036200903010026
GOAPhAR: An Integrative Discovery Tool for Annotation, Pathway Analysis
Abstract
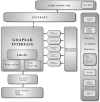
We have developed the web based tool GOAPhAR (Gene Ontology, Annotations and Pathways for Array Research), that integrates information from disparate sources regarding gene annotations, protein annotations, identifiers associated with probe sets, functional pathways, protein interactions, Gene Ontology, publicly available microarray datasets and tools for statistically validating clusters in microarray data. Genes of interest can be input as Affymetrix probe identifiers, Genbank, or Unigene identifiers for human, mouse or rat genomes. Results are provided in a user friendly interface with hyperlinks to the sources of information.
Figures
References
-
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. - PubMed
-
- Lockhart DJ. Expression monitoring by hybridization to high-density oligonucleotide array. Nat Biotechnol. 1996;14(13):1675–80. - PubMed
-
- Bolstad BM. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. - PubMed
-
- Riva A. Comments on selected fundamental aspects of microarray analysis. Comput Biol Chem. 2005;29(5):319–36. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources