An NMR database for simulations of membrane dynamics
- PMID: 21134351
- PMCID: PMC5176272
- DOI: 10.1016/j.bbamem.2010.11.027
An NMR database for simulations of membrane dynamics
Abstract
Computational methods are powerful in capturing the results of experimental studies in terms of force fields that both explain and predict biological structures. Validation of molecular simulations requires comparison with experimental data to test and confirm computational predictions. Here we report a comprehensive database of NMR results for membrane phospholipids with interpretations intended to be accessible by non-NMR specialists. Experimental ¹³C-¹H and ²H NMR segmental order parameters (S(CH) or S(CD)) and spin-lattice (Zeeman) relaxation times (T(1Z)) are summarized in convenient tabular form for various saturated, unsaturated, and biological membrane phospholipids. Segmental order parameters give direct information about bilayer structural properties, including the area per lipid and volumetric hydrocarbon thickness. In addition, relaxation rates provide complementary information about molecular dynamics. Particular attention is paid to the magnetic field dependence (frequency dispersion) of the NMR relaxation rates in terms of various simplified power laws. Model-free reduction of the T(1Z) studies in terms of a power-law formalism shows that the relaxation rates for saturated phosphatidylcholines follow a single frequency-dispersive trend within the MHz regime. We show how analytical models can guide the continued development of atomistic and coarse-grained force fields. Our interpretation suggests that lipid diffusion and collective order fluctuations are implicitly governed by the viscoelastic nature of the liquid-crystalline ensemble. Collective bilayer excitations are emergent over mesoscopic length scales that fall between the molecular and bilayer dimensions, and are important for lipid organization and lipid-protein interactions. Future conceptual advances and theoretical reductions will foster understanding of biomembrane structural dynamics through a synergy of NMR measurements and molecular simulations.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures

 ) 15.04 MHz, (
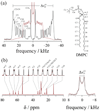
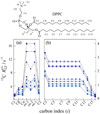
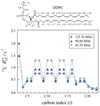
) 15.04 MHz, ( ) 20.00 MHz, (◆) 25.15 MHz, (▼) 45.29 MHz, (▲) 90.80 MHz, (●) 125.76 MHz, and (■) 150.84 MHz. (a) The glycerol backbone and choline headgroup are resolved in natural-abundance 13C NMR in the 50–80 ppm region and exhibit a pronounced dispersion of the relaxation rates. (b) Acyl chain segments are observed in the 0–40 ppm fingerprint region of the high-resolution 13C NMR spectrum. The molecular structure of DPPC and assignments corresponding to the carbon index (i) are shown. Data taken in part from Ref. [96].
) 20.00 MHz, (◆) 25.15 MHz, (▼) 45.29 MHz, (▲) 90.80 MHz, (●) 125.76 MHz, and (■) 150.84 MHz. (a) The glycerol backbone and choline headgroup are resolved in natural-abundance 13C NMR in the 50–80 ppm region and exhibit a pronounced dispersion of the relaxation rates. (b) Acyl chain segments are observed in the 0–40 ppm fingerprint region of the high-resolution 13C NMR spectrum. The molecular structure of DPPC and assignments corresponding to the carbon index (i) are shown. Data taken in part from Ref. [96].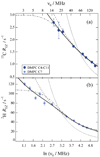
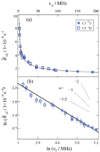
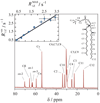
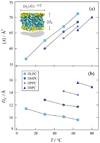
 ) 2H NMR are fit by a single power-law function (——) with n = −1/2 consistent with a membrane deformation model. (b) Double-logarithmic plots of scaled relaxation rate dispersion with comparative fitting to alternative power-law frequency scalings as indicated. Power-law exponents are shown for n = −2, −1, and −1/2 corresponding to molecular diffusion, flexible surface (smectic deformation), and membrane deformation (nematic-like) models. Data taken in part from Refs. [46,96].
) 2H NMR are fit by a single power-law function (——) with n = −1/2 consistent with a membrane deformation model. (b) Double-logarithmic plots of scaled relaxation rate dispersion with comparative fitting to alternative power-law frequency scalings as indicated. Power-law exponents are shown for n = −2, −1, and −1/2 corresponding to molecular diffusion, flexible surface (smectic deformation), and membrane deformation (nematic-like) models. Data taken in part from Refs. [46,96].
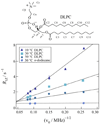
References
-
- Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q. Rev. Biophys. 1977;10:353–418. - PubMed
-
- Griffin RG. Solid state nuclear magnetic resonance of lipid bilayers. Method Enzymol. 1981;72:108–174. - PubMed
-
- Davis JH. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta. 1983;737:117–171. - PubMed
-
- Bloom M, Evans E, Mouritsen OG. Physical properties of the fluid lipid-bilayer component of cell membranes: A perspective. Quart. Rev. Biophys. 1991;24:293–397. - PubMed
-
- Brown MF. Membrane structure and dynamics studied with NMR spectroscopy. In: Merz K Jr, Roux B, editors. Biological Membranes. A Molecular Perspective from Computation and Experiment. Basel: Birkhäuser; 1996. pp. 175–252.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

