HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid
- PMID: 21134384
- PMCID: PMC3046808
- DOI: 10.1016/j.jmb.2010.11.051
HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid
Abstract
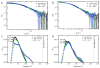
The retroviral Gag polyprotein mediates viral assembly. The Gag protein has been shown to interact with other Gag proteins, with the viral RNA, and with the cell membrane during the assembly process. Intrinsically disordered regions linking ordered domains make characterization of the protein structure difficult. Through small-angle scattering and molecular modeling, we have previously shown that monomeric human immunodeficiency virus type 1 (HIV-1) Gag protein in solution adopts compact conformations. However, cryo-electron microscopic analysis of immature virions shows that in these particles, HIV-1 Gag protein molecules are rod shaped. These differing results imply that large changes in Gag conformation are possible and may be required for viral formation. By recapitulating key interactions in the assembly process and characterizing the Gag protein using neutron scattering, we have identified interactions capable of reversibly extending the Gag protein. In addition, we demonstrate advanced applications of neutron reflectivity in resolving Gag conformations on a membrane. Several kinds of evidence show that basic residues found on the distal N- and C-terminal domains enable both ends of Gag to bind to either membranes or nucleic acid. These results, together with other published observations, suggest that simultaneous interactions of an HIV-1 Gag molecule with all three components (protein, nucleic acid, and membrane) are required for full extension of the protein.
Published by Elsevier Ltd.
Figures



References
-
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview: 1997. pp. 263–334. - PubMed
-
- Fuller SD, Wilk T, Gowen BE, Kräusslich HG, Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

