SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons
- PMID: 21135118
- PMCID: PMC3028638
- DOI: 10.1128/MCB.01117-10
SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons
Abstract
SR proteins are well known to promote exon inclusion in regulated splicing through exonic splicing enhancers. SR proteins have also been reported to cause exon skipping, but little is known about the mechanism. We previously characterized SRSF1 (SF2/ASF)-dependent exon skipping of the CaMKIIδ gene during heart remodeling. By using mouse embryo fibroblasts derived from conditional SR protein knockout mice, we now show that SR protein-induced exon skipping depends on their prevalent actions on a flanking constitutive exon and requires collaboration of more than one SR protein. These findings, coupled with other established rules for SR proteins, provide a theoretical framework to understand the complex effect of SR protein-regulated splicing in mammalian cells. We further demonstrate that heart-specific CaMKIIδ splicing can be reconstituted in fibroblasts by downregulating SR proteins and upregulating a RBFOX protein and that SR protein overexpression impairs regulated CaMKIIδ splicing and neuronal differentiation in P19 cells, illustrating that SR protein-dependent exon skipping may constitute a key strategy for synergism with other splicing regulators in establishing tissue-specific alternative splicing critical for cell differentiation programs.
Figures



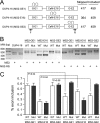



References
-
- Anko, M. L., L. Morales, I. Henry, A. Beyer, and K. M. Neugebauer. 2010. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat. Struct. Mol. Biol. 17:962-970. - PubMed
-
- Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
