Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model
- PMID: 21135205
- PMCID: PMC3009777
- DOI: 10.1073/pnas.1009845107
Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model
Abstract
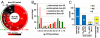
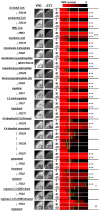
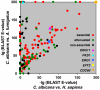
One potentially rich source of possible targets for antifungal therapy are those Candida albicans genes deemed essential for growth under the standard culture (i.e., in vitro) conditions; however, these genes are largely unexplored as drug targets because essential genes are not experimentally amenable to conventional gene deletion and virulence studies. Using tetracycline-regulatable promoter-based conditional mutants, we investigated a murine model of candidiasis in which repressing essential genes in the host was achieved. By adding doxycycline to the drinking water starting 3 days prior to (dox - 3D) or 2 days post (dox + 2D) infection, the phenotypic consequences of temporal gene inactivation were assessed by monitoring animal survival and fungal burden in prophylaxis and acute infection settings. Of 177 selected conditional shut-off strains tested, the virulence of 102 was blocked under both repressing conditions, suggesting that the corresponding genes are essential for growth and survival in a murine host across early and established infection periods. Among these genes were those previously identified as antifungal drug targets (i.e., FKS1, ERG1, and ERG11), verifying that this methodology can be used to validate potential new targets. We also identify genes either conditionally essential or dispensable for in vitro growth but required for survival and virulence, including those in late stage ergosterol synthesis, or early steps in fatty acid or riboflavin biosynthesis. This study evaluates the role of essential genes with respect to pathogen virulence in a large-scale, systems biology context, and provides a general method for gene target validation and for uncovering unexpected antimicrobial targets.
Conflict of interest statement
Conflict of interest statement: Some of the authors are employees of Merck & Co., Inc., as stated in the affiliations, and potentially own stock and/or holdings in the company.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

