Determinants of the specificity of rotavirus interactions with the alpha2beta1 integrin
- PMID: 21138834
- PMCID: PMC3057834
- DOI: 10.1074/jbc.M110.142992
Determinants of the specificity of rotavirus interactions with the alpha2beta1 integrin
Abstract
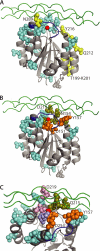
The human α2β1 integrin binds collagen and acts as a cellular receptor for rotaviruses and human echovirus 1. These ligands require the inserted (I) domain within the α2 subunit of α2β1 for binding. Previous studies have identified the binding sites for collagen and echovirus 1 in the α2 I domain. We used CHO cells expressing mutated α2β1 to identify amino acids involved in binding to human and animal rotaviruses. Residues where mutation affected rotavirus binding were located in several exposed loops and adjacent regions of the α2 I domain. Binding by all rotaviruses was eliminated by mutations in the activation-responsive αC-α6 and αF helices. This is a novel feature that distinguishes rotavirus from other α2β1 ligands. Mutation of residues that co-ordinate the metal ion (Ser-153, Thr-221, and Glu-256 in α2 and Asp-130 in β1) and nearby amino acids (Ser-154, Gln-215, and Asp-219) also inhibited rotavirus binding. The importance of most of these residues was greatest for binding by human rotaviruses. These mutations inhibit collagen binding to α2β1 (apart from Glu-256) but do not affect echovirus binding. Overall, residues where mutation affected both rotavirus and collagen recognition are located at one side of the metal ion-dependent adhesion site, whereas those important for collagen alone cluster nearby. Mutations eliminating rotavirus and echovirus binding are distinct, consistent with the respective preference of these viruses for activated or inactive α2β1. In contrast, rotavirus and collagen utilize activated α2β1 and show an overlap in α2β1 residues important for binding.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

