Structure of Importin13-Ubc9 complex: nuclear import and release of a key regulator of sumoylation
- PMID: 21139563
- PMCID: PMC3025465
- DOI: 10.1038/emboj.2010.320
Structure of Importin13-Ubc9 complex: nuclear import and release of a key regulator of sumoylation
Abstract
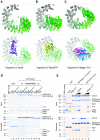
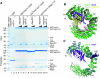
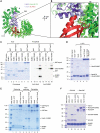
Importin13 (Imp13) is an unusual β-karyopherin that is able to both import and export cargoes in and out of the nucleus. In the cytoplasm, Imp13 associates with different cargoes such as Mago-Y14 and Ubc9, and facilitates their import into the nucleus where RanGTP binding promotes the release of the cargo. In this study, we present the 2.8 Å resolution crystal structure of Imp13 in complex with the SUMO E2-conjugating enzyme, Ubc9. The structure shows an uncommon mode of cargo-karyopherin recognition with Ubc9 binding at the N-terminal portion of Imp13, occupying the entire RanGTP-binding site. Comparison of the Imp13-Ubc9 complex with Imp13-Mago-Y14 shows the remarkable plasticity of Imp13, whose conformation changes from a closed ring to an open superhelix when bound to the two different cargoes. The structure also shows that the binding mode is compatible with the sumoylated states of Ubc9. Indeed, we find that Imp13 is able to bind sumoylated Ubc9 in vitro and suppresses autosumoylation activity in the complex.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Andrade MA, Petosa C, O'Donoghue SI, Müller CW, Bork P (2001) Comparison of ARM and HEAT protein repeats. J Mol Biol 309: 1–18 - PubMed
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 - PubMed
-
- Bono F, Cook AG, Grunwald M, Ebert J, Conti E (2010) Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of Importin 13. Mol Cell 37: 211–222 - PubMed
-
- Bono F, Ebert J, Lorentzen E, Conti E (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

