HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens
- PMID: 21139978
- PMCID: PMC2994760
HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens
Abstract
Purpose: An interlocking network of transcription factors, RNA binding proteins, and miRNAs globally regulates gene expression and alternative splicing throughout development, and ensures the coordinated mutually exclusive expression of non-neural and neuronal forms of these factors during neurogenesis. Striking similarities between lens fiber cell and neuron cell morphology led us to determine if these factors are also used in the lens. HuR and polypyrimidine tract binding protein (PTB) have been described as 'global regulators' of RNA alternative splicing, stability, and translation in non-neuronal (including ectodermal) tissues examined to date in diverse species, and REST/NRSF (RE-1 Silencing Transcription Factor/Neuron Restrictive Silencing Factor) represses>2,000 neuronal genes in all non-neuronal tissues examined to date, but has not included the lens. During neurogenesis these factors are replaced by what has been considered neuron-specific HuB/C/D, nPTB, and alternatively spliced REST (REST4), which work with miR-124 to activate this battery of genes, comprehensively reprogram neuronal alternative splicing, and maintain their exclusive expression in post-mitotic neurons.
Methods: Immunoprecipitation, western blot, immunofluorescence, and immunohistochemistry were used to determine the expression and distribution of proteins in mouse and rat lenses. Mobility shift assays were used to examine lenses for REST/NRSF DNA binding activity, and RT-PCR, DNA sequencing, and northern blots were used to identify RNA expression and alternative splicing events in lenses from mouse, rat, and goldfish (N. crassa).
Results: We demonstrated that REST, HuR, and PTB proteins are expressed predominantly in epithelial cells in mouse and rat lenses, and showed these factors are also replaced by the predominant expression of REST4, HuB/C/D and nPTB in post-mitotic fiber cells, together with miR-124 expression in vertebrate lenses. REST-regulated gene products were found to be restricted to fiber cells where REST is decreased. These findings predicted nPTB- and HuB/C/D-dependent splicing reactions can also occur in lenses, and we showed Neuronal C-src and Type 1 Neurofibromatosis 1 splicing as well as calcitonin gene related peptide (CGRP) and neural cell adhesion molecule (NCAM-180) alternative transcripts in lenses. Transgenic mice with increased HuD in lens also showed increased growth associated protein 43 (GAP43) and Ca++/Calmodulin dependent kinase IIα (CamKIIα) HuD target gene expression in the lens, similar to brain.
Conclusions: The present study provides the first evidence this fundamental set of regulatory factors, previously considered to have a unique role in governing neurogenesis are also used in the lens, and raises questions about the origins of these developmental factors and mechanisms in lens and neuronal cells that also have a basic role in determining the neuronal phenotype.
Figures


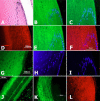

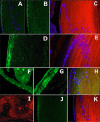





References
-
- Alberts B. Molecular Biology of the Cell. Fourth ed. London: Garland Science; 2007.
-
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–42. - PubMed
-
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell. 2009;36:1007–17. - PubMed
-
- Jones FS, Meech R. Knockout of REST/NRSF shows that the protein is a potent repressor of neuronally expressed genes in non-neural tissues. Bioessays. 1999;21:372–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
