Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly
- PMID: 21142019
- PMCID: PMC4355582
- DOI: 10.1021/bi101005r
Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly
Abstract
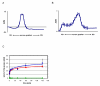
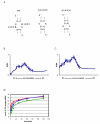
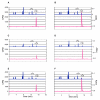
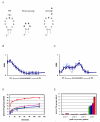
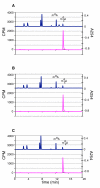
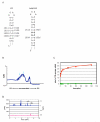
KsgA is an rRNA methyltransferase important to the process of small subunit biogenesis in bacteria. It is ubiquitously found in all life including archaea and eukarya, where the enzyme is referred to as Dim1. Despite the emergence of considerable data addressing KsgA function over the last several years, details pertaining to RNA recognition are limited, in part because the most accessible substrate for in vitro studies of KsgA is the 900000 Da 30S ribosomal subunit. To overcome challenges imposed by size and complexity, we adapted recently reported techniques to construct in vivo assembled mutant 30S subunits suitable for use in in vitro methyltransferase assays. Using this approach, numerous 16S rRNA mutants were constructed and tested. Our observations indicate that the 790 loop of helix 24 plays an important role in overall catalysis by KsgA. Moreover, the length of helix 45 also is important to catalysis. In both cases loss of catalytic function occurred without an increase in the production of N(6)-methyladenosine, a likely indication that there was no critical reduction in binding strength. Both sets of observations support a "proximity" mechanism of KsgA function. We also report that several of the mutants constructed failed to assemble properly into 30S subunits, while some others did so with reduced efficiency. Therefore, the same technique of generating mutant 30S subunits can be used to study ribosome biogenesis on the whole.
Figures
References
-
- Helser TL, Davies JE, Dahlberg JE. Change in Methylation of 16S Ribosomal RNA Associated with Mutation to Kasugamycin Resistance in Escherichia Coli. Nat. New Biol. 1971;233:12–14. - PubMed
-
- Poldermans B, Roza L, Van Knippenberg PH. Studies on the Function of Two Adjacent N6, N6-Dimethyladenosines Near the 3′ End of 16 S Ribosomal RNA of Escherichia Coli. III. Purification and Properties of the Methylating Enzyme and Methylase-30 S Interactions. J. Biol. Chem. 1979;254:9094–9100. - PubMed
-
- Van Buul CP, Damm JB, Van Knippenberg PH. Kasugamycin Resistant Mutants of Bacillus Stearothermophilus Lacking the Enzyme for the Methylation of Two Adjacent Adenosines in 16S Ribosomal RNA. Mol. Gen. Genet. 1983;189:475–478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

