Thermodynamic characterization of the binding interaction between the histone demethylase LSD1/KDM1 and CoREST
- PMID: 21142040
- PMCID: PMC3604701
- DOI: 10.1021/bi101776t
Thermodynamic characterization of the binding interaction between the histone demethylase LSD1/KDM1 and CoREST
Abstract
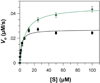
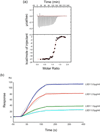
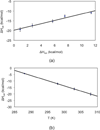
Flavin-dependent histone demethylases catalyze the posttranslational oxidative demethylation of mono- and dimethylated lysine residues, producing formaldehyde and hydrogen peroxide in addition to the corresponding demethylated protein. In vivo, histone demethylase LSD1 (KDM1; BCH110) is a component of the multiprotein complex that includes histone deacetylases (HDAC 1 and 2) and the scaffolding protein CoREST. Although little is known about the affinities of or the structural basis for the interaction between CoREST and HDACs, the structure of CoREST(286-482) bound to an α-helical coiled-coil tower domain within LSD1 has recently been reported. Given the significance of CoREST in directing demethylation to specific nucleosomal substrates, insight into the molecular basis of the interaction between CoREST and LSD1 may suggest a new means of inhibiting LSD1 activity by misdirecting the enzyme away from nucleosomal substrates. Toward this end, isothermal titration calorimetry studies were conducted to determine the affinity and thermodynamic parameters characterizing the binding interaction between LSD1 and CoREST(286-482). The proteins tightly interact in a 1:1 stoichiometry with a dissociation constant (K(d)) of 15.9 ± 2.07 nM, and their binding interaction is characterized by a favorable enthalpic contribution near room temperature with a smaller entropic penalty at pH 7.4. Additionally, one proton is transferred from the buffer to the heterodimeric complex at pH 7.4. From the temperature dependence of the enthalpy change of interaction, a constant-pressure heat capacity change (ΔC(p)) of the interaction was determined to be -0.80 ± 0.01 kcal mol(-1) K(-1). Notably, structure-driven truncation of CoREST revealed that the central binding determinant lies within the segment of residues 293-380, also known as the CoREST "linker" region, which is a central isolated helix that interacts with the LSD1 coiled-coil tower domain to create a triple-helical bundle. Thermodynamic parameters obtained from the binding between LSD1 and the linker region of CoREST are similar to those obtained from the interaction between LSD1 and CoREST(286-482). These results provide a framework for understanding the molecular basis of protein-protein interactions that govern nucleosomal demethylation.
Figures



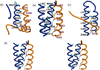
Similar articles
-
Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase.Mol Cell. 2006 Aug 4;23(3):377-87. doi: 10.1016/j.molcel.2006.07.012. Mol Cell. 2006. PMID: 16885027
-
Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex.Nucleic Acids Res. 2015 May 26;43(10):4868-80. doi: 10.1093/nar/gkv388. Epub 2015 Apr 27. Nucleic Acids Res. 2015. PMID: 25916846 Free PMC article.
-
LSD1/CoREST is an allosteric nanoscale clamp regulated by H3-histone-tail molecular recognition.Proc Natl Acad Sci U S A. 2012 Jul 31;109(31):12509-14. doi: 10.1073/pnas.1207892109. Epub 2012 Jul 16. Proc Natl Acad Sci U S A. 2012. PMID: 22802671 Free PMC article.
-
Targeting the LSD1/KDM1 Family of Lysine Demethylases in Cancer and Other Human Diseases.Adv Exp Med Biol. 2023;1433:15-49. doi: 10.1007/978-3-031-38176-8_2. Adv Exp Med Biol. 2023. PMID: 37751134 Review.
-
LSD1: more than demethylation of histone lysine residues.Exp Mol Med. 2020 Dec;52(12):1936-1947. doi: 10.1038/s12276-020-00542-2. Epub 2020 Dec 14. Exp Mol Med. 2020. PMID: 33318631 Free PMC article. Review.
Cited by
-
G-quadruplex RNA binding and recognition by the lysine-specific histone demethylase-1 enzyme.RNA. 2016 Aug;22(8):1250-60. doi: 10.1261/rna.057265.116. Epub 2016 Jun 8. RNA. 2016. PMID: 27277658 Free PMC article.
-
Lysine-Specific Demethylase 1 Inhibitors: A Comprehensive Review Utilizing Computer-Aided Drug Design Technologies.Molecules. 2024 Jan 22;29(2):550. doi: 10.3390/molecules29020550. Molecules. 2024. PMID: 38276629 Free PMC article. Review.
-
A selective phenelzine analogue inhibitor of histone demethylase LSD1.ACS Chem Biol. 2014 Jun 20;9(6):1284-93. doi: 10.1021/cb500018s. Epub 2014 Apr 7. ACS Chem Biol. 2014. PMID: 24707965 Free PMC article.
-
A rationally-designed chimeric KDM1A/KDM1B histone demethylase tower domain deletion mutant retaining enzymatic activity.FEBS Lett. 2015 Aug 19;589(18):2340-6. doi: 10.1016/j.febslet.2015.07.028. Epub 2015 Jul 29. FEBS Lett. 2015. PMID: 26226427 Free PMC article.
-
Evolution of lysine-specific demethylase 1 and REST corepressor gene families and their molecular interaction.Commun Biol. 2023 Dec 14;6(1):1267. doi: 10.1038/s42003-023-05652-x. Commun Biol. 2023. PMID: 38097664 Free PMC article.
References
-
- Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev. 2003;13:127–135. - PubMed
-
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. - PubMed
-
- Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. - PubMed
-
- Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

