The SAM-responsive S(MK) box is a reversible riboswitch
- PMID: 21143313
- PMCID: PMC3064258
- DOI: 10.1111/j.1365-2958.2010.07410.x
The SAM-responsive S(MK) box is a reversible riboswitch
Abstract
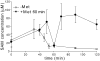
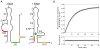
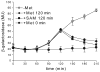
The S(MK) (SAM-III) box is an S-adenosylmethionine (SAM)-responsive riboswitch found in the 5' untranslated region of metK genes, encoding SAM synthetase, in many members of the Lactobacillales. SAM binding causes a structural rearrangement in the RNA that sequesters the Shine-Dalgarno (SD) sequence by pairing with a complementary anti-SD (ASD) sequence; sequestration of the SD sequence inhibits binding of the 30S ribosomal subunit and prevents translation initiation. We observed a slight increase in the half-life of the metK transcript in vivo when Enterococcus faecalis cells were depleted for SAM, but no significant change in overall transcript abundance, consistent with the model that this riboswitch regulates at the level of translation initiation. The half-life of the SAM-S(MK) box RNA complex in vitro is shorter than that of the metK transcript in vivo, raising the possibility of reversible binding of SAM. We used a fluorescence assay to directly visualize reversible switching between the SAM-free and SAM-bound conformations. We propose that the S(MK) box riboswitch can make multiple SAM-dependent regulatory decisions during the lifetime of the transcript in vivo, acting as a reversible switch that allows the cell to respond rapidly to fluctuations in SAM pools by modulating expression of the SAM synthetase gene.
© 2010 Blackwell Publishing Ltd.
Figures
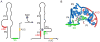

Similar articles
-
S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA.Proc Natl Acad Sci U S A. 2007 Mar 20;104(12):4876-80. doi: 10.1073/pnas.0609956104. Epub 2007 Mar 9. Proc Natl Acad Sci U S A. 2007. PMID: 17360376 Free PMC article.
-
Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.Nat Struct Mol Biol. 2008 Oct;15(10):1076-83. doi: 10.1038/nsmb.1494. Epub 2008 Sep 21. Nat Struct Mol Biol. 2008. PMID: 18806797 Free PMC article.
-
The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase.Nat Struct Mol Biol. 2006 Mar;13(3):226-33. doi: 10.1038/nsmb1059. Epub 2006 Feb 19. Nat Struct Mol Biol. 2006. PMID: 16491091
-
Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis.J Hepatol. 2013 Oct;59(4):830-41. doi: 10.1016/j.jhep.2013.04.031. Epub 2013 May 7. J Hepatol. 2013. PMID: 23665184 Review.
-
Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine.Biochem Cell Biol. 2008 Apr;86(2):157-68. doi: 10.1139/O08-008. Biochem Cell Biol. 2008. PMID: 18443629 Review.
Cited by
-
Themes and variations in riboswitch structure and function.Biochim Biophys Acta. 2014 Oct;1839(10):908-918. doi: 10.1016/j.bbagrm.2014.02.012. Epub 2014 Feb 28. Biochim Biophys Acta. 2014. PMID: 24583553 Free PMC article. Review.
-
TaRTLEt: Transcriptionally-active Riboswitch Tracer Leveraging Edge deTection.PeerJ. 2025 May 26;13:e19418. doi: 10.7717/peerj.19418. eCollection 2025. PeerJ. 2025. PMID: 40444283 Free PMC article.
-
NusG-dependent RNA polymerase pausing is a common feature of riboswitch regulatory mechanisms.Nucleic Acids Res. 2024 Nov 27;52(21):12945-12960. doi: 10.1093/nar/gkae981. Nucleic Acids Res. 2024. PMID: 39494516 Free PMC article.
-
Riboswitch control of induction of aminoglycoside resistance acetyl and adenyl-transferases.RNA Biol. 2013 Aug;10(8):1266-73. doi: 10.4161/rna.25757. Epub 2013 Jul 15. RNA Biol. 2013. PMID: 23880830 Free PMC article.
-
Fingerprinting Tertiary Structure in Complex RNAs Using Single-Molecule Correlated Chemical Probing.Biochemistry. 2024 Oct 15;63(20):2648-2657. doi: 10.1021/acs.biochem.4c00343. Epub 2024 Oct 3. Biochemistry. 2024. PMID: 39359229
References
-
- Edwards TE, Klein DJ, Ferre-D’Amare AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol. 2007;17:273–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

